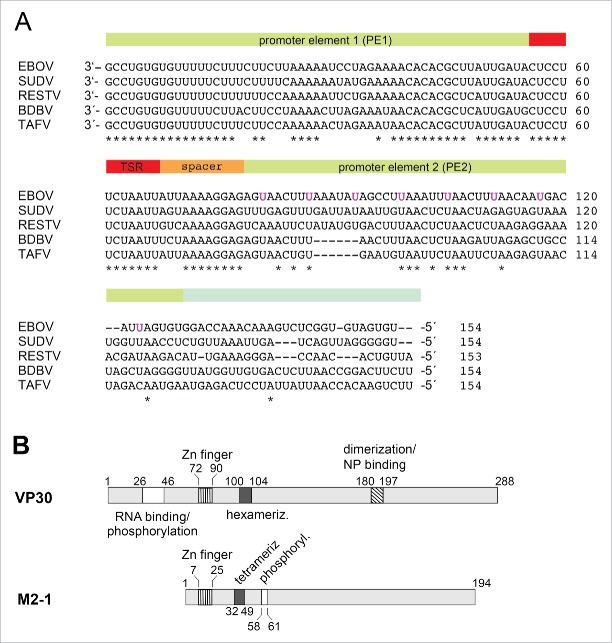
Figure 1.
(A) Alignment of the 3′-leader region (∼154 nucleotides) of different Ebola species; the bipartite replication promoter is indicated by the green-shaded and the transcription start region (TSR) by the red-shaded bar regions above the sequences; 5 while the 3′-terminal ∼ 78 nt of the genomic (−) RNA are strongly conserved, the different EBOV sequences become more diverse further upstream (nt 79–154). The uracils of the 8 UN5 hexamers of the PE2 element of Zaire ebolavirus5 are marked in magenta. EBOV: Zaire ebolavirus; BDBV: Bundibugyo ebolavirus; SUDV: Sudan ebolavirus; RESTV: Reston ebolavirus; TAFV: Tai Forest ebolavirus. (B) Schematic comparison of Ebola VP30 and related M2–1 transcription factors of Paramyxoviridae. In VP30 proteins, the N-terminal domain, which is absent in M2–1 proteins, harbors the region of amino acids (aa) 26–46 involved in RNA binding and phosphorylation (white box; 12,30 the zinc finger domain (hatched with vertical lines, aa 72–90;31),the hexamerization motif (black box, aa 100–104; 32) and the region mediating VP30 dimerization and binding to NP (diagonally hatched box;17). For M2–1, the corresponding Zn finger domain and the core domain harboring the tetramerization helix and the phosphorylation targets S58 and S61 are depicted.23 For more details, see Fig. S7.