Figure 6.
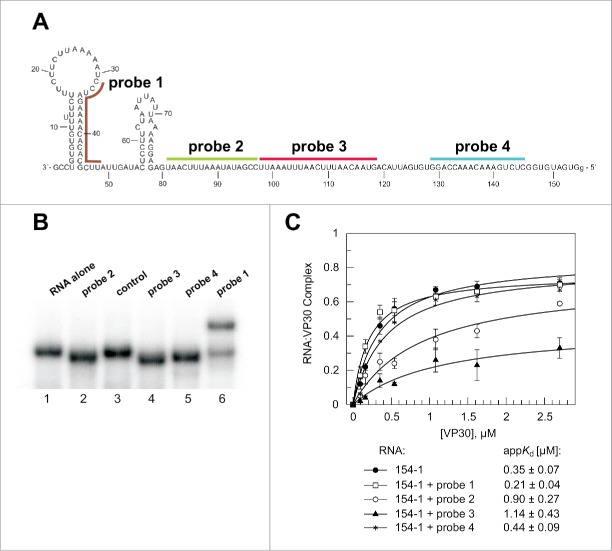
Hybridization of DNA probes to the genomic RNA 154–1. (A) Secondary structure (as experimentally defined, see Fig. 2) of the genomic RNA and position of DNA probes that were annealed to the RNA before VP30:RNA gel shift assays. (B) Native PAA gel to verify stable annealing of the different probes to the RNA substrate. The RNA (154–1) without any DNA oligonucleotide bound (RNA alone) showed the same mobility as the RNA preincubated with a better: non-complementary DNA oligonucleotide (control). The DNA probes complementary to single-stranded RNA region resulted in accelerated mobility of the RNA (probes 2, 3 and 4), whereas probe 1 disrupting a double-stranded RNA regions led to retarded RNA mobility, also suggesting that hybridization was not complete. (C) VP30:RNA binding curves and derived appKd values (± SD) of the different preannealed RNA:DNA probe complexes. Each curve is based on at least 3 independent experiments (error bars are standard deviations of the mean); curves were fitted according to a one-ligand-binding-site model.