Figure 2.
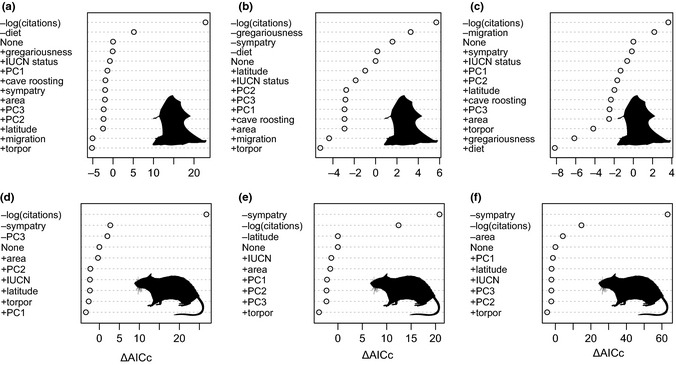
Ranking of host trait variables from the GLS models by ΔAICc: the change in AICc values when each variable is individually added (+) or removed (−) from the best model for (a) the number of viruses identified in a bat species (best model: number of viruses ∼ log(citations) + diet), (b) the degree of a bat species in the viral sharing network (best model: degree ∼ log(citations) + gregariousness + sympatry + diet), (c) the betweenness of a bat species in the viral sharing network (best model: betweenness ∼ log(citations) + migration), (d) the number of viruses identified in a rodent species (best model: number of viruses ∼ log(citations) + sympatry + PC3), (e) the degree of a rodent species in the viral sharing network (best model: degree ∼ log(citations) + sympatry + latitude) and (f) the betweenness of a rodent species in the viral sharing network (best model: betweenness ∼ log(citations) + sympatry + area).
