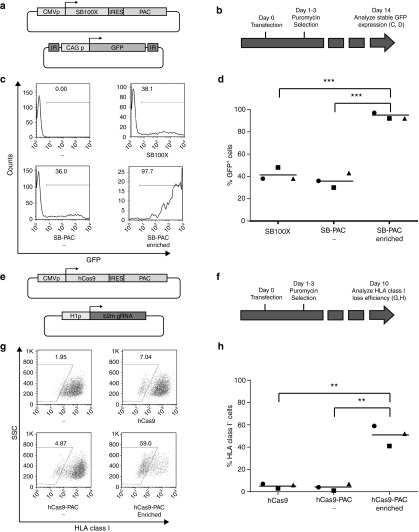
Figure 3.
Drug selection based enrichment of SB and hCas9 gene-modified cells. (a) Vectors used to evaluate the effect of puromycin selection on stable SB100X transposition. CAG p, chicken β-actin promoter with CMV enhancer; IRES, hepatitis C virus internal ribosome entry site; PAC, puromycin N-acetyl-transferase cassette. (b) Schematic experimental time line. HeLa cells were transfected and, after 24 hours, cultured in the presence of puromycin for 48 hours. 14 days after transfection, stable transposition was assessed by flow cytometry. (c) Flow cytometric analysis of the effect of early puromycin selection on stable GFP transposition. Puromycin-selected cells (bottom right plot) become almost uniformly GFP positive. (d) Summary of three independent SB gene transfer enrichment experiments. In all cases, early puromycin selection leads to very high stable GFP expression (***P < 0.001). (e) Vectors used to evaluate the effect of puromycin selection on hCas9-mediated genome editing. hCas9, humanized Cas9; H1p, DNA polymerase III promoter H1; b2m sgRNA, β2M-specific guiding RNA. (f) Schematic experimental time line. HeLa cells were transfected and, after 24 hours, cultured in the presence of puromycin for 48 hours. Fourteen days after transfection, stable HLA class I loss was evaluated by flow cytometry. (g) Flow cytometric analysis of the effect of early puromycin selection on stable HLA class I loss. Puromycin-treated cells (bottom right plot) are highly enriched for HLA class I-negative cells. (h) Summary of three independent hCas9 genome editing enrichment experiments. In all cases, early puromycin selection greatly increases the percentage of cells with stable HLA class I loss (**P value < 0.01).