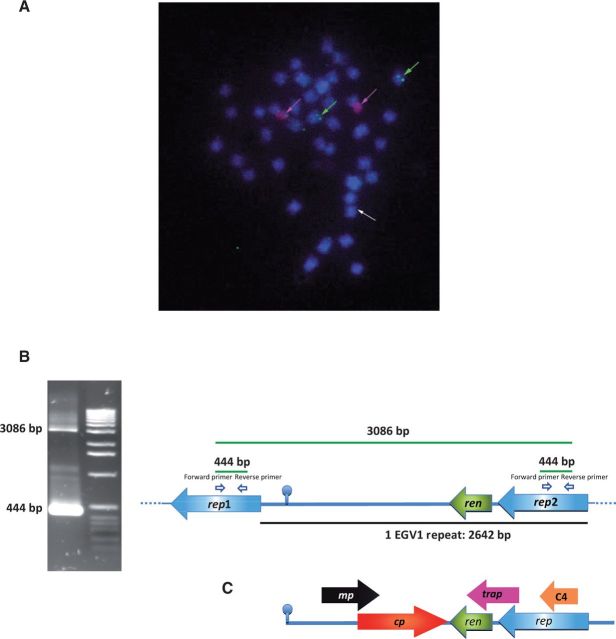
Figure 1.
(A) FISH on D. alata chromosomes with a 2.6 kb EGV1 probe from D. alata (detected in green) and a rDNA 45 S probe used as control (detected in red). Chromosomes are counterstained with DAPI, in blue. Scale bar = 5 µm. The green arrows indicate four hybridization signals potentially corresponding to the presence of tandem repeats of EGV located on both chromatids of two chromosome pairs. The white arrows indicate faint signals potentially corresponding to the presence of copies of the rep gene scattered around the genome of D. alata. (B) Long template PCRs enabled the amplification of two DNA fragments (444 bp and 3,086 bp) from D. alata acc. 313. Sequencing of 444 and 3,086 fragments revealed a partial tandem repeat genomic organization with two partial rep genes, a ren gene, and an intergenic region containing a GC-rich stem carrying a nonanucleotide loop TAATATTAC. The size of one repeat is 2,642 bp. (C) Representation of a typical linearized begomovirus genome in the virion sense orientation starting from the origin of replication (mp, movement protein; cp, capsid protein; ren, replication enhancer; trap, transactivator protein; rep, replication-associated protein; C4 ORF has been shown to suppress transcriptional gene silencing).