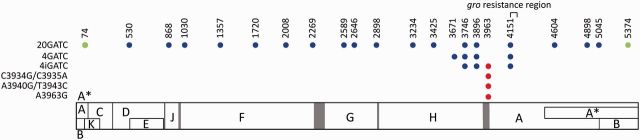
Figure 1.
ϕX174 genetic map and location of the GATC sequence motifs introduced in this study. Open reading frames are represented by rectangles (B, K, and E are in different reading frames), and gray bars indicate intergenic regions. Each GATC is represented by a dot, and its position is indicated on top. GATC motifs that were synonymous in all reading frames are indicated in blue, whereas those producing amino acid replacements in at least one frame are shown in green (A74C produces a K494Q replacement in gene A and is synonymous in gene K; T5374G produces a V465G replacement in gene A and is synonymous in gene B). Mutations falling at intergenic regions are shown in red. The phage has circular DNA but is represented linearly for convenience, where by convention the first position corresponds to the last nucleotide of the unique PstI site.