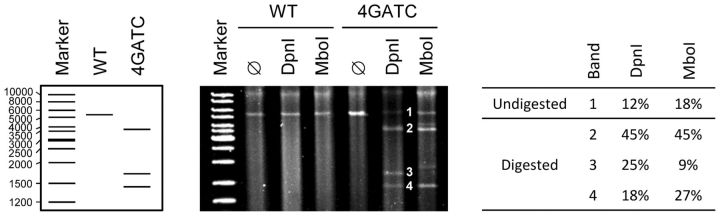
Figure 3.
Restriction fragment analysis of the ϕX174 replicative dsDNA. Phage dsDNA was purified by standard miniprep as described in the Materials and Methods section and linearized with XhoI (ø), which recognizes a unique site at position 162, with XhoI and DpnI to cleave methylated or hemi-methylated GATCs, or with XhoI and MboI to cleave non-methylated GATCs. Expected (left) and observed (center) restriction fragments for the WT and 4GATC phage dsDNA are shown. Lower size fragments (<300 bp, Fig. 1) were expected but could not be visualized because the amount of input DNA was low. The smear in lanes containing the purified phage DNA probably results from degradation of host DNA. The contrast of the gel image was enhanced to help visualize bands. Right: percent abundance of each DpnI and MboI restriction band (1–4). Band 1 in the DpnI lane indicates the non-methylated DNA fraction (i.e., none of the four GATC motifs was methylated), whereas in the MboI lane, this same band indicates the fully methylated fraction (i.e., the four GATC motifs were methylated). Bands were quantified as detailed in the Materials and Methods section using the raw gel image with no contrast enhancement.