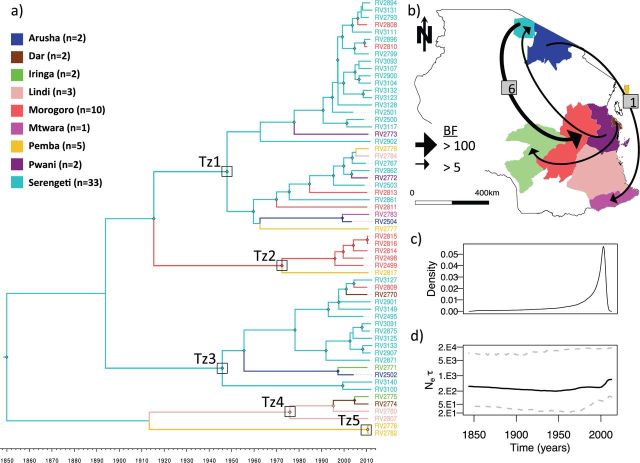
Figure 2.
Regional phylogeography among sixty rabies virus whole-genome sequences sampled in Tanzania from 2003 to 2012: (a) an MCC tree with branches coloured according to the most probable posterior location of its descendent node inferred by discrete-state phylogeographic reconstruction in BEAST. Five major phylogenetic groups (Tz1-5) are annotated on the tree and node symbols indicate node posterior support ≥0.9. (b) The four most significant dispersal pathways indicated by BF results from a BSSVS procedure in BEAST with the median number of transitions estimated by Markov jump counts indicated in cases where posterior support for a transition was >0.7. (c) Markov jump densities for total number of transitions through time. (d) Bayesian Skyline plot showing Neτ, the product of the effective population size (Ne), and the generation time in years (τ) through time.