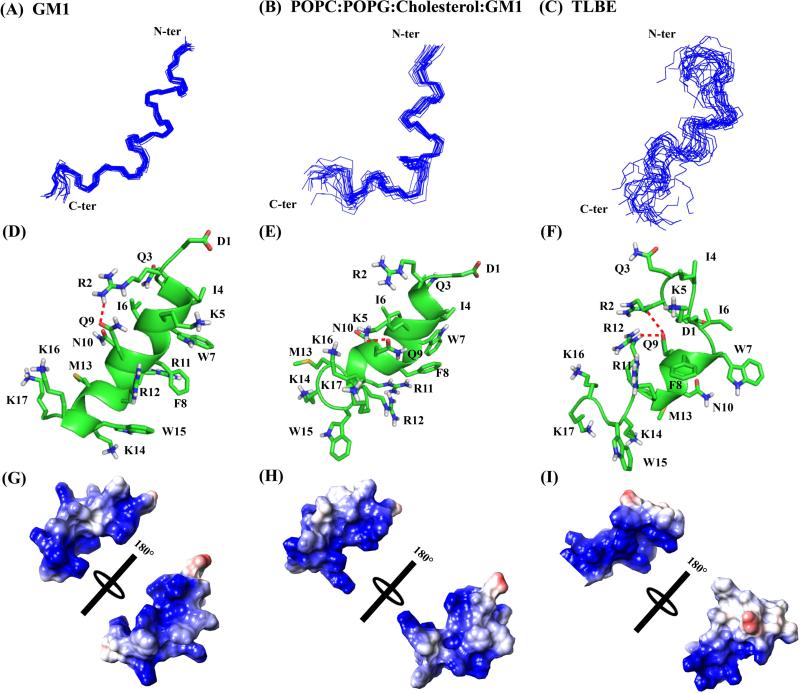
Figure 6.
Three-dimensional solution NMR structures of DK17 peptide in various membrane models in 10 mM Na2HPO4 buffer, pH 4.5. Superposition of backbone atoms (N, Cα, C’) of the 20 lowest energy structures of DK17 bound to GM1 (A), POPC:POPG:Cholesterol:GM1 (B), and TLBE (C) LUVs. Cartoon representations of side chain orientation of 2D-NMR derived structure of DK17 in GM1 (D), POPC:POPG:Cholesterol:GM1 (E) and TLBE (F) LUVs showing different residues. The red dotted lines indicate either electrostatic interaction or hydrogen bonding between the side chain residues. Electrostatic surface potential map for DK17 in GM1 (D), POPC:POPG:Cholesterol:GM1 (E) and TLBE (F) LUVs showing charged residues.