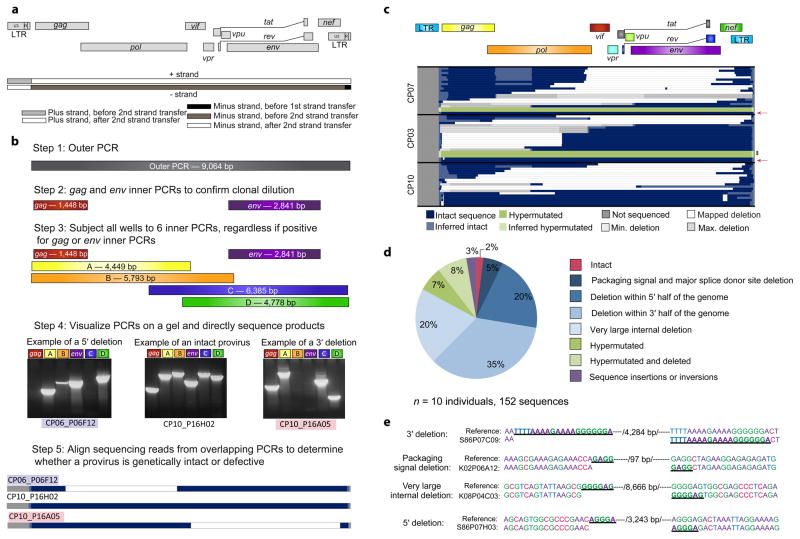
Figure 1.
Proviral sequences in chronically-treated subjects are highly defective. (a) Schematic of strand transfer events during reverse transcription relative to the HIV-1 genome. Deletions occur during minus strand synthesis before the second strand transfer event by the same copy choice mechanism that leads to recombination25. (b) Unbiased full genome sequencing strategy based on mechanistic considerations in (a) to capture the majority of defective proviruses. Bands from representative proviruses with a 5′ deletion, an intact sequence, and a 3′ deletion are shown. (c) Maps of proviral clones from three representative subjects treated during the chronic phase of infection. Each horizontal bar represents an individual clone identified through full genome sequencing. In cases where a deletion could not be precisely mapped due to a deletion encompassing multiple forward or reverse primer binding sites, the possible maximum and minimum deletions sizes are plotted (in grey). If sequencing data shows a mapped deletion that removes primer-binding sites for other amplicons, the resulting missing sequence was inferred to be present (light blue or green). A pink arrow denotes full-length, genetically intact sequences. Black asterisks indicate likely full-length hypermutated proviruses that were not fully sequenced due to extreme hypermutation. See Supplementary Fig. 1 for maps of proviruses from seven additional subjects. (d) Summary of 152 proviral sequences. (e) Short repeats (underlined) identified on both ends of the deletion junctions are consistent with a copy choice mechanism of recombination resulting in deletion of the intervening sequence and one homology region (/number of basepairs deleted/).