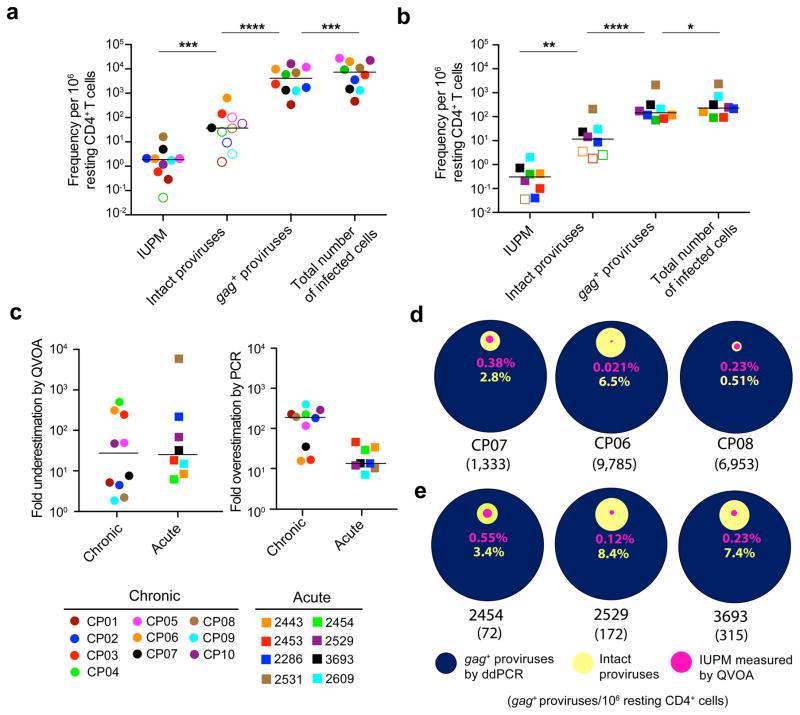
Figure 4.
Current assays significantly underestimate or overestimate the size of the latent reservoir. (a,b) Comparison of different reservoir measurements in resting CD4+ T cells from subjects starting ART during chronic (a) or acute (b) infection. The frequency of infected cells was measured by QVOA, which detects cells that release infectious virus after one round of T cell activation and is reported as the number of infectious units per million resting CD4+ T cells (IUPM). For individuals in whom the IUPM was below the limit of detection, the median posterior estimate of infection frequency was plotted instead (open symbols). The predicted total number of infected cells was calculated for each subject by correcting the ddPCR result (gag+ proviruses) for the fraction of proviruses with deletions in gag. The frequency of cells with intact proviruses was calculated as the frequency of infected cells times the fraction of intact proviruses estimated for each subject using an empirical Bayesian model. Open symbols indicate subjects in which no intact proviruses were detected. Horizontal bars indicate median values. Statistical significance was determined using a two-tailed paired t-test and the variance was similar between groups. *P < 0.05; ** P < 0.01; *** P < 0.001; **** P < 0.0001. (c) Underestimation of the latent reservoir size by the QVOA was calculated by dividing the number of intact proviruses by the QVOA result for each subject. Overestimation of the latent reservoir size by DNA PCR was calculated by dividing the gag+ provirus PCR values by the number of intact proviruses. Horizontal bars indicate median values. (d,e) Comparison of infected cell frequencies as determined by QVOA (pink), analysis of intact proviruses (yellow), and ddPCR for gag+ proviral DNA (blue) for three representative CP- (d) and AP- treated (e) subjects. All values are plotted as a percentage of the frequency of cells with gag+ proviral DNA. The number of gag+ proviruses per million resting CD4+ T cells as measured by ddPCR is indicated in parenthesis for each subject. For additional subject comparison plots, see Supplementary Fig. 7d,e.