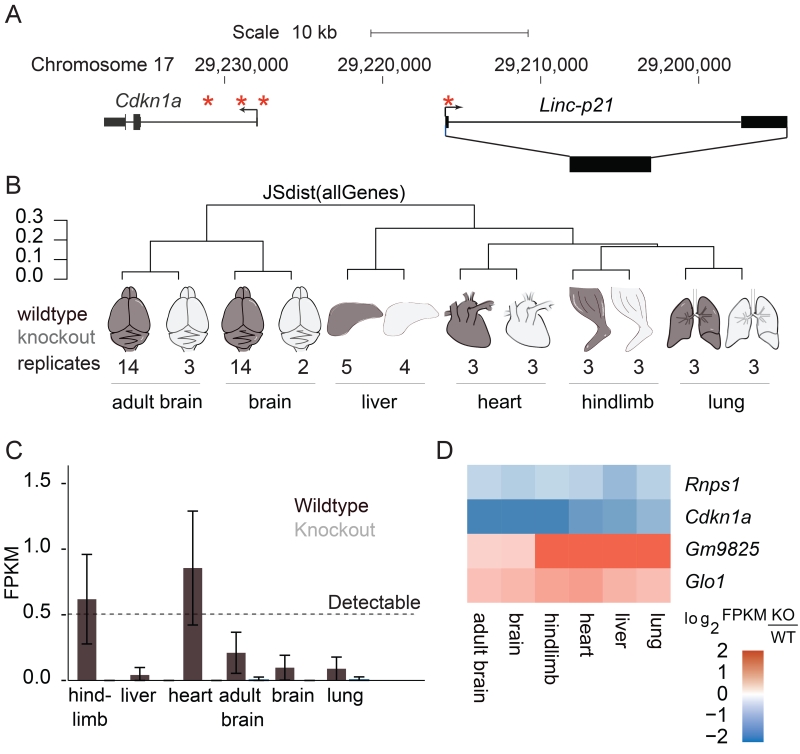
Figure 1. Linc-p21 in vivo deletion overview.
(A) Linc-p21/Cdkn1a locus on mouse chromosome 17. Asterisks indicate known p53 binding sites. (B) Dendrogram showing the type and number of samples sequenced and the Jenson-Shanon distance, a measure of total transcriptome similarity, between their expression profiles. Wildtype shown in black, knockout shown in grey. (C) Average Linc-p21 expression profile in each tissue. Error bars represent 95% confidence interval. (D) Heatmap depicting expression of genes significantly differentially expressed in every tissue. Log2 fold-change was calculated using the average FPKMs for all replicates (KO/WT). See also Figure S1.