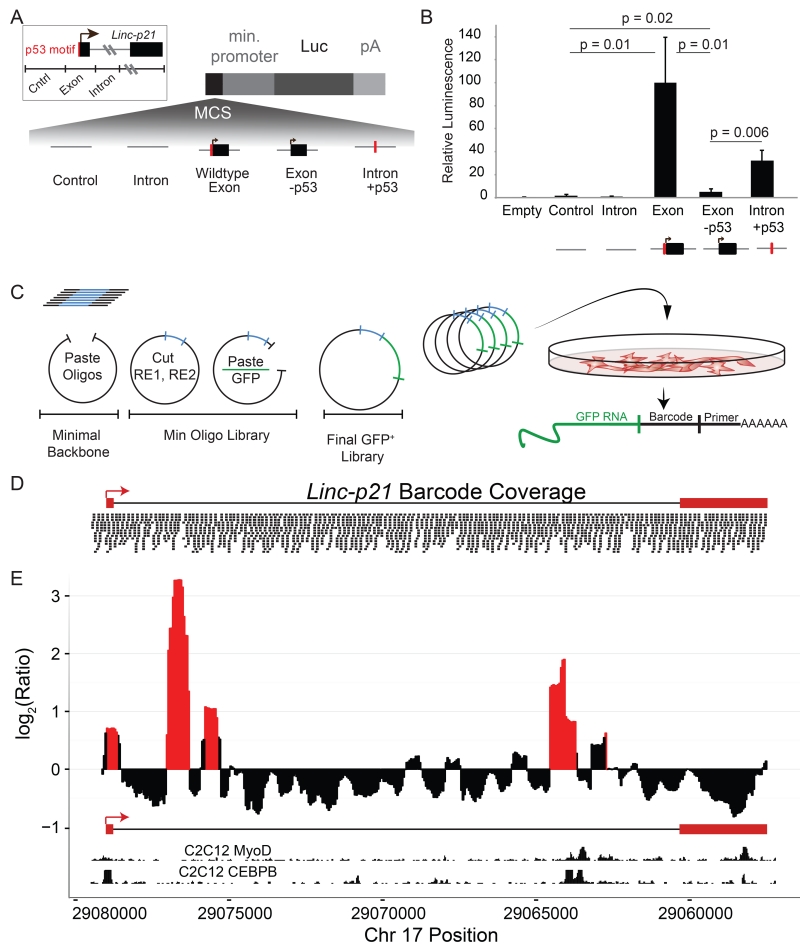
Figure 4. MPRA of the entire Linc-p21 locus reveals enhancer activity.
(A) Experimental design for luciferase reporter assay using an intergenic control and regions from the Linc-p21 promoter and exon 1 (“exon”), intron 1 (“intron”), exon with mutated p53 binding sequence (“exon -p53”) and intron with p53 binding sequence (“intron+p53”). Each region was cloned into a luciferase reporter construct. P53 motif indicated in red. (B) Relative luminescence for each construct, normalized against the signal from exon fragment and averaged across triplicate samples. P values were calculated using unpaired one-tailed t-tests. Error bars represent standard error of the mean. (C) Massively Parallel Reporter Assay (MPRA) experimental design: oligos were synthesized, subcloned into a minimal backbone, opened by enzymatic digestion and re-ligated with a GFP cDNA insert. Pooled constructs were transfected into C2C12 cells in triplicate and libraries were made from GFP+ RNA. (D) Coverage of final pooled GFP+ vector library across the Linc-p21 locus and promoter (assessed by high throughput sequencing). (E) MPRA signal across the Linc-p21 locus. Y-axis represents the log2 ratio of normalized RNA to control signal per base (averaged in 500bp sliding windows every 50bp). Significance (p<0.01) is calculated by comparing this signal to 1000 random shuffles of the input data. Significant peaks are shown in red. Inlayed tracks are CEBPB and MyoD ChIP-seq signals from UCSC genome browser. See also Figure S4.