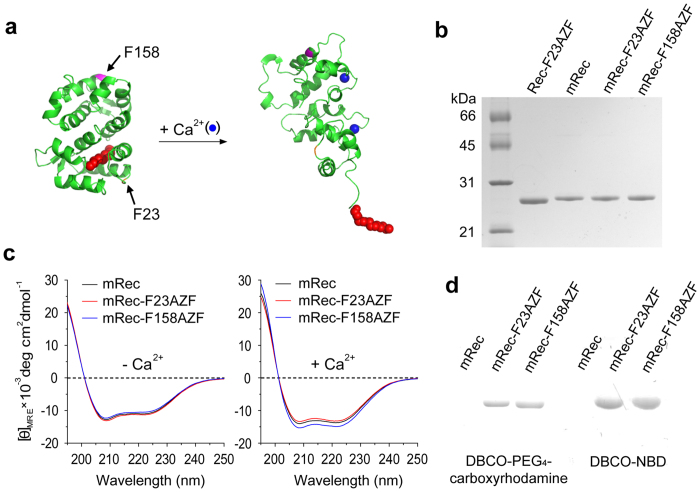
Figure 1. Genetic incorporation of the non-natural amino acid AZF into two selected positions of recoverin.
(a) Three dimensional structures of recoverin in its Ca2+-free (PDB code: 1IKU; left) and Ca2+-bound (PDB code: 1JSA; right) forms. Sites selected for mutation are marked with arrows. The N-myristoyl chain is represented by red spheres. (b) SDS-PAGE of purified recoverin and variants: Lane 1, non-myristoylated Rec-F23AZF; Lane 2, myristoylated recoverin; Lane 3, mRec-F23AZF; Lane 4, mRec-F158AZF. (c) Far-UV CD spectra of recoverin and variants in the absence or presence of 1 mM Ca2+. (d) In-gel fluorescence of recoverin and variants reacted with DBCO-PEG4-carboxyrhodamine (left) or DBCO-NBD (right): Lane 1, mRec; Lane 2, mRec-F23AZF; Lane 3, mRec-F158AZF. The labeled protein bands run at 24 kDa as shown with the mCherry reference in Supplementary Fig. 4.