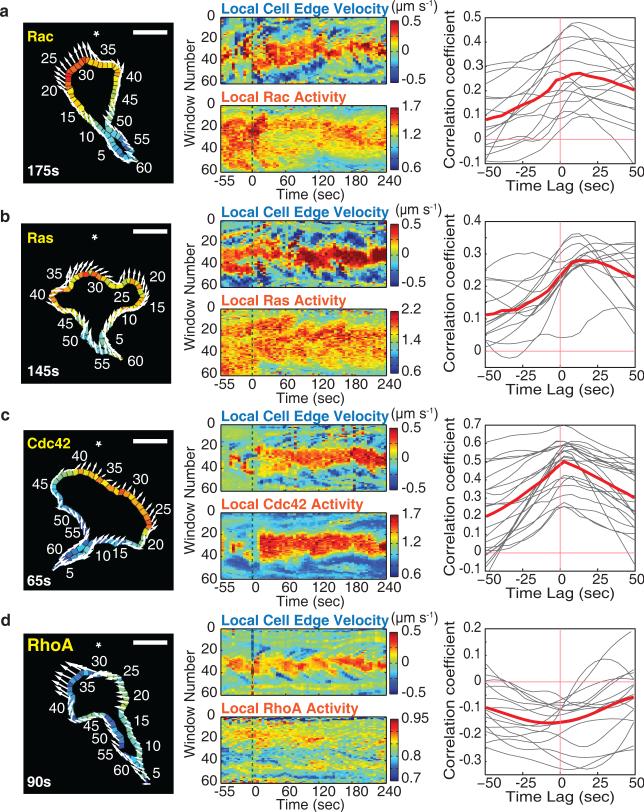
Figure 7.
Cdc42 and opposing RhoA activities correlate spatiotemporally with local protrusion during cell migration. (a-d) Left, example image of local GTPase activities measured in 60 segmented regions along the cell edge. Arrows indicate the movement of the corresponding edge segments between sequential images. The direction of the chemoattractant gradient is indicated with a *. Time after stimulation is marked. Scale bar is 10μm. Middle, spatial and temporal activity maps of edge segment protrusion dynamics against local GTPase activities. Color bars indicate protrusion-retraction dynamics and activation level of FRET biosensors. Green dotted lines mark the start of gradient generation by photorelease. Right, temporal cross-correlation analysis between edge protrusion and the respective GTPase activities averaged over all sampling segments as a function of time offset. Negative time lag values indicate GTPase activity preceding protrusion activity. The gray curves are data for individual cells, and the red curves represent the mean for all cells measured. n=15 (Rac), n=14 (Ras), n=22 (Cdc42), and n=15 (RhoA) cells.