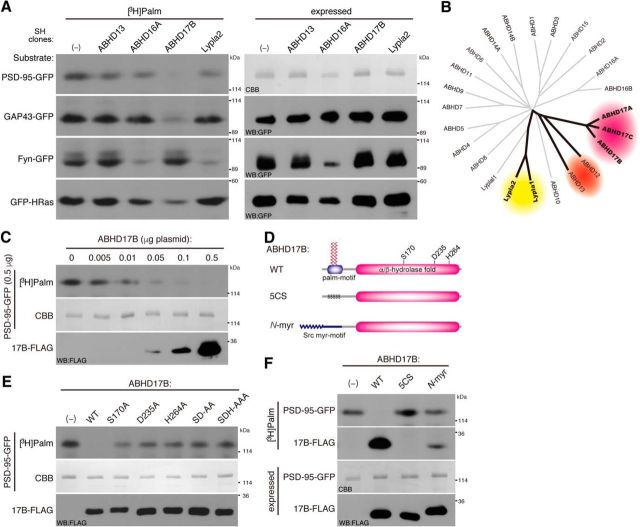
Figure 2.
Enzymatic characterization of ABHD17. A, Substrate specificity of ABHD proteins and Lypla2 was tested in HEK293T cells as in Figure 1. B, Phylogenetic tree of mouse ABHD genes. The tree is based on alignment of the α/β hydrolase fold domains. Putative subgroups for PSD-95 depalmitoylating enzyme candidates are shown by ovals in different colors. C, Increasing amounts of cotransfected ABHD17B plasmid caused PSD-95 depalmitoylation (0.5 μg plasmid) in a dose-dependent manner. D, ABHD17B mutant constructs. ABHD17B–5CS (C10,11,14,15,18S), palmitoylation-deficient ABHD17B; N-myr, myristoylated ABHD17B in which the N-terminal palmitoylation motif (aa 1–20) was replaced with myristoylation motif of Src (aa 1–20). The catalytic triad (S170/D235/H264) is indicated. E, F, Mutagenesis of ABHD17B showed that the catalytic triad (S170/D235/H264; E) and the N-terminal palmitoylation motif (F) were essential for the depalmitoylating activity of ABHD17B to PSD-95. Note that wild-type ABHD17B, but not ABHD17B–5CS, was highly palmitoylated. The weak [3H]palmitate labeling of N-myr ABHD17B might reflect N-palmitoylation, as N-myristoyl transferase can use palmitic acid as a substrate in vitro, albeit at a reduced efficiency compared with myristate (Kishore et al., 1993; Martin et al., 2011). SD-AA, ABHD17B-S170A/D235A; SDH-AAA, ABHD17B-S170A/D235A/H264A.