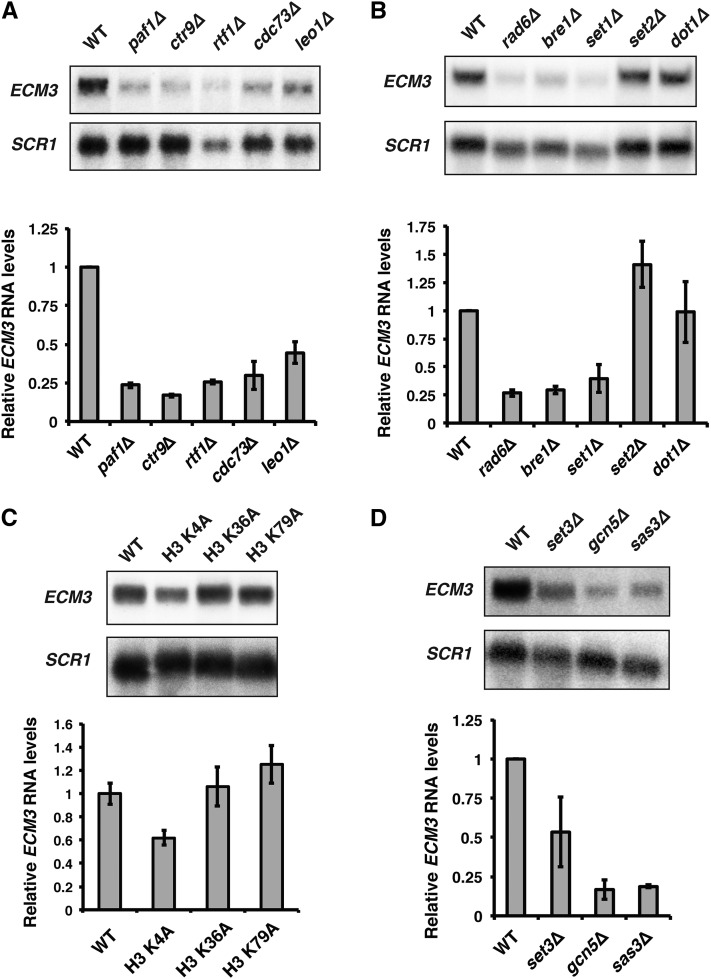
Figure 2.
The Paf1 complex and methylation of H3 K4 positively regulate ECM3 expression. (A) Representative northern blot analysis of ECM3 transcript levels in a wild-type strain (FY4) or strains lacking one of the five subunits of the Paf1 complex, either paf1∆ (YJ807), ctr9∆ (KY2170), rtf1∆ (YJ788), cdc73∆ (KY2171), or leo1∆ (KY1805). (B) Representative northern blot analysis of ECM3 transcript levels in a wild-type strain (FY4) and strains where the genes encoding histone modifiers that work in concert with the Paf1 complex have been deleted (rad6∆, KY1712; bre1∆, KY1713; set1∆, KY2720; set2∆, KY2723; and dot1∆, KY2725). (C) Representative northern blot analysis of ECM3 transcript levels in a wild-type control strain, lacking one copy of the genes for H3 and H4 (JDY86), and derivatives of JDY86 in which the only copy of the H3-H4 genes encodes the indicated amino acid substitution in H3. (D) Representative northern blot analysis of ECM3 transcript levels in strains lacking a subunit of the Set3 HDAC complex (set3∆, KY2782), the SAGA HAT complex (gcn5∆, KY1743), or the NuA3 HAT complex (sas3∆, ECY394) compared to wild-type levels (YJ1125). Quantitation below shows the average ECM3 mRNA levels relative to WT (set to 1) from at least three biological replicates. Error bars represent the SEM. SCR1 serves as a loading control. mRNA, messenger RNA; WT, wild-type.