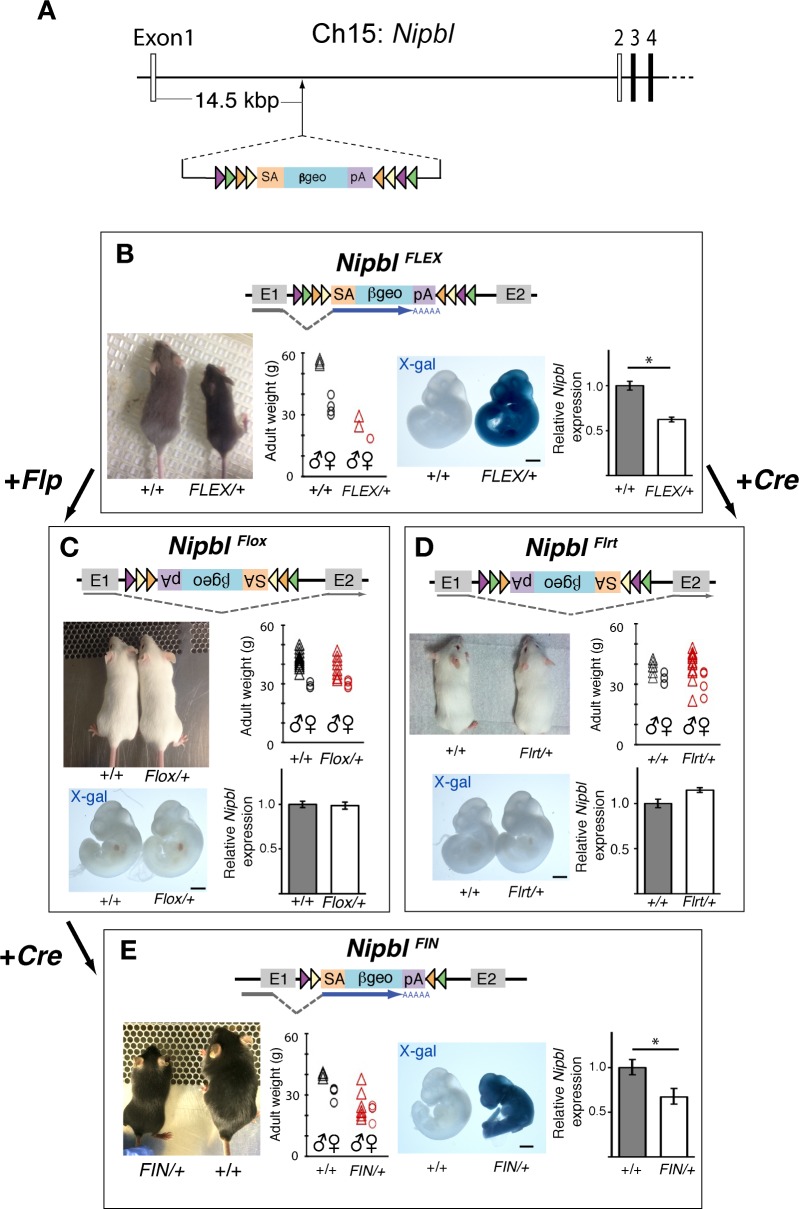
Fig 2. FLEX alleles allow successive toggling between mutant and wildtype genotypes and phenotypes.
A. Schematic of EUCE313f02 (NipblFLEX) allele from which the NipblFLEX/+ mouse line and allelic series are derived. The rsFlp-Rosa-βgeo cassette is inserted 14.5 kbp downstream of Nipbl Exon 1 on Chromosome 15. B. In the NipblFLEX allele, the splice acceptor (SA) in the cassette traps Nipbl expression, resulting in termination of Nipbl expression after exon 1 and expression of the β-geo reporter for the trapped null allele. Adult NipblFLEX/+ mice are smaller than wildtype littermates: Image is of 4-wk-old male littermates. Scatter plot shows weights of 12-wk-old NipblFLEX/+ mice (red, n = 3: 1 female, 2 males) and wildtype littermates (black, n = 8: 4 females, 4 males) from 3 litters. Ubiquitous expression of β-geo was detected by X-gal staining in E10.5 NipblFLEX/+ embryos. Histogram shows mean ± SEM of relative Nipbl expression, assessed by qRT-PCR, in kidneys of E17.5 NipblFLEX/+ (n = 8) and wildtype littermates (n = 6); asterisk: p < 0.05 by Student’s t test. C. Mating NipblFLEX/+ mice with mice carrying universal Flp recombinase inverts the SA-βgeo-pA at heterotypic recognition targets (frt and F3 sites) and simultaneously excises cognate recognition sites, resulting in progeny carrying the NipblFlox/+ allele. Inversion allows normal splicing between the endogenous Nipbl splice sites (Exon 1 to Exon 2), thereby yielding a phenotypically wildtype allele. NipblFlox/+ mice are similar to wildtype littermates in size: Image is of 3-wk-old male littermates; scatter plot shows weights of 11-wk-old NipblFlox/+ mice (red, n = 18: 4 female; 14 male) compared to wildtype littermates (black, n = 19: 4 female; 15 male) from 5 litters. Expression of β-geo is not detected by X-gal staining in E10.5 NipblFlox/+ embryos. Histogram shows qRT-PCR analysis of relative Nipbl expression in brain tissue of E17.5 in NipblFlox/+ (n = 8) versus wildtype littermates (n = 7), plotted as in B; p > 0.05, Student’s t test. D. Mating NipblFLEX/+ mice with mice carrying a universal Cre recombinase causes recombination of the NipblFLEX allele (at LoxP and lox5171 recognition sites), resulting in progeny carrying the NipblFlrt allele. NipblFlrt/+mice are phenotypically wildtype: Image is of male NipblFlrt/+ and wildtype littermates at 3 wk of age showing no apparent difference in body size. Scatter plot shows weights of 12-wk-old NipblFlrt/+ mice (red, n = 19: 6 female; 13 male) and wildtype littermates (black, n = 10: 3 female; 7 male) from 3 litters. Expression of β-geo was not detected by X-gal staining in E10.5 NipblFlrt/+ embryos. qRT-PCR results show relative Nipbl expression in kidneys of E17.5 NipblFlrt/+ (n = 6) compared to wildtype littermates (n = 6), plotted as in B; p > 0.05 by Student’s t test. E. Cre-mediated recombination of mice carrying the NipblFlox allele, obtained by crossing NipblFlox/+ mice with Nanog-Cre hemizygous mice, results in re-inversion of the SA-βgeo-pA cassette and re-trapping of Nipbl expression. Resulting progeny (NipblFIN/+ mice) are phenotypically mutant, and survive poorly, with only 13 NipblFIN/+ mice (4%) surviving to weaning age out of 315 total pups born (significantly less than the expected 25% survival, p < 0.001 by Chi-square analysis). Adult NipblFIN/+ mice are smaller than wildtype littermates: Image is of 6-wk old males; scatter plot shows weights of 8-wk-old NipblFIN/+ mice (red, n = 11: 4 females; 7 males) compared to wildtype littermates (black, n = 7: 3 females; 4 males) from 16 litters. Ubiquitous expression of β-geo is detected by X-gal staining. qRT-PCR results show reduced Nipbl expression in brains of E17.5 NipblFIN/+ (n = 7) compared to wildtype littermates (n = 6), plotted as in B; asterisk: p < 0.05, Student’s t test. Scale bars = 1 mm for all panels. Frt (purple triangles), F3 (green triangles), loxP (orange triangles) and lox5171 (yellow triangles); SA, splice acceptor; β-geo, β-galactosidase/neomycin phosphotransferase fusion gene; pA, bovine growth hormone polyadenylation sequence.