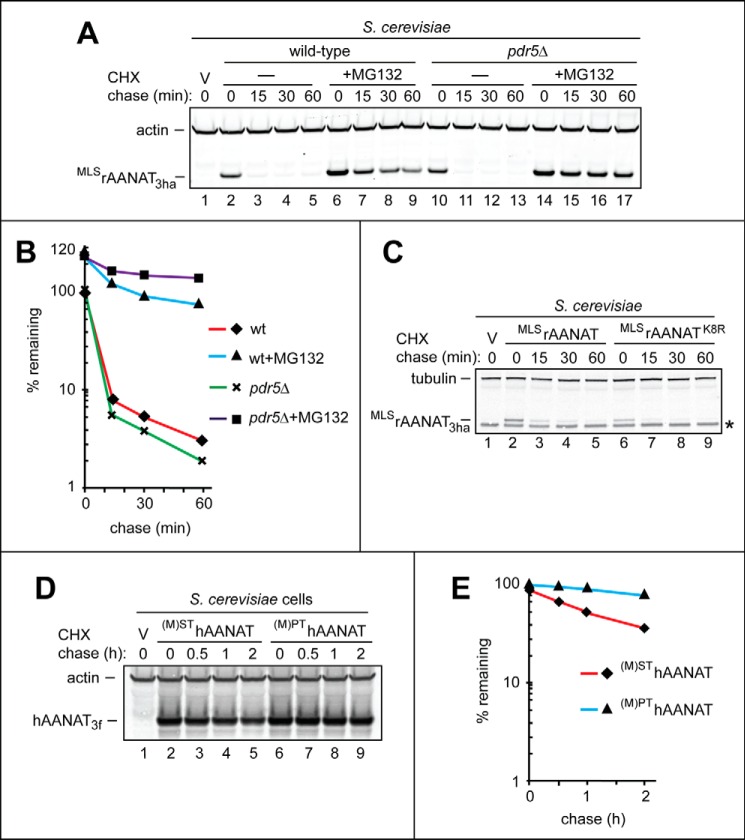
FIGURE 4.
Degradation assays with a proteasome inhibitor and AANAT mutants in S. cerevisiae. A, lane 1, wild-type S. cerevisiae were transformed with vector (V) alone (control). Lanes 2–5, CHX-chase was performed at 30 °C for the indicated times in wild-type S. cerevisiae with the wild-type rat MLSrAANAT3ha. Lanes 6–9, same as in lanes 2–5, but the CHX-chase was in the presence of the MG132 proteasome inhibitor (see “Experimental Procedures”). Lanes 10–13, same as in lanes 2–5 but in pdr5Δ S. cerevisiae lacking the efflux pump Pdr5. Lanes 14–17, same as in lanes 10–13 but in the presence of MG132. The bands of actin and MLSrAANAT3ha are indicated on the left. B, quantification of data in A. C, CHX-chases were performed in wild-type S. cerevisiae with the wild-type rat MLSrAANAT3ha (lanes 2–5) and with its Lys-8 → Arg mutant MLSrAANAT3haK8R (see “Results”). The bands of tubulin and AANAT are indicated on the left. An asterisk on the right denotes a band of protein that cross-reacted with anti-HA antibody (the band is also present in lane 1, the vector-only control). D, lane 1, wild-type S. cerevisiae were transformed with vector (V) alone (control). Lanes 2–5, CHX-chase was performed at 30 °C for the indicated times in wild-type S. cerevisiae with the wild-type human (M)SThAANAT3f. Lanes 6–9, same as in lanes 2–5 but with the mutant human (M)PThAANAT3f. The bands of actin and AANAT are indicated on the left. E, quantification of data in A. All quantified CHX-chase assays were carried out at least three times and yielded results within 10% of the data shown.