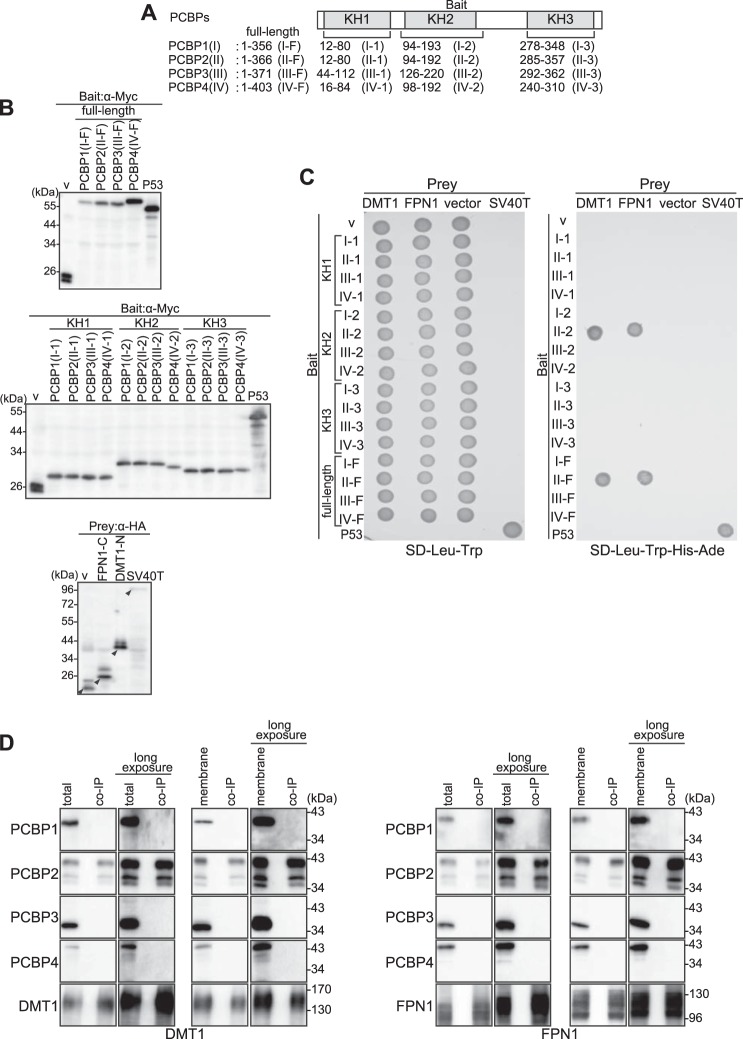
FIGURE 2.
DMT1 and FPN1 can bind to PCBP2 but not PCBP1, PCBP3, or PCBP4 in yeast or mammalian cells. A, analysis of the interactions of DMT1 or FPN1 with the four PCBPs using a yeast two-hybrid assay. PCBP1 (amino acids 12–80 (I-1), 94–193 (I-2), 278–348 (I-3), or 1–356 (I-F)); PCBP2 (amino acids 12–80 (II-1), 94–192 (II-2), 285–357 (II-3), or 1–366 (II-F)); PCBP3 (amino acids 44–112 (III-1), 126–220 (III-2), 292–362 (III-3), or 1–371 (III-F)); and PCBP4 (amino acids 16–84 (IV-1), 98–192 (IV-2), 240–310 (IV-3), or (1–403 (IV-F)) were cloned into a vector containing the GAL4 DNA-binding domain (bait vector). Vectors containing the GAL4 DNA-activating domain fused to the N-terminal cytoplasmic region of DMT1 or the C-terminal cytoplasmic region of FPN1 were used as prey. B, confirmation of bait and prey expression. Yeast cells were harvested, and whole protein was prepared using the method described under “Experimental Procedures.” Bait protein expression was detected by using anti-Myc mAb, and prey was detected by anti-HA mAb. The arrowheads indicate the appropriate molecular weight of each recombinant protein. Results are typical experiments of at least three performed. v, vector. C, analysis of the interactions of DMT1 or FPN1 with PCBPs. After mating yeast expressing the bait protein with yeast expressing the prey protein, yeast cells were grown on both SD/-Trp/-Leu and SD/-Trp/-Leu/-His/-Ade selection media and incubated for 48 h at 30 °C. The vector alone served as a negative control, and the well characterized p53 and SV40-T interaction served as a positive control, as suggested by the manufacturer. Similar results were obtained from at least three independent experiments. D, analysis of the interactions of DMT1 or FPN1 with PCBP1–4 using a co-immunoprecipitation assay. HEp-2 cells were transfected with FPN1-GFP or DMT1A-I-GFP and incubated for 48 h at 37 °C. After incubation, total protein was extracted in TNE buffer and assessed by immunoblotting using the appropriate anti-PCBP1, -2, -3, or -4 or GFP Ab (left lane). The membrane fraction was prepared using the method described under “Experimental Procedures.” Using either the total protein extract or the membrane fraction, co-immunoprecipitation analyses (co-IP) were performed. The precipitates were analyzed by immunoblotting with anti-PCBP1 pAb, anti-PCBP2 mAb, anti-PCBP3 pAb, anti-PCBP4 pAb, and anti-GFP pAb (right lane). Similar results were obtained from at least three independent experiments.