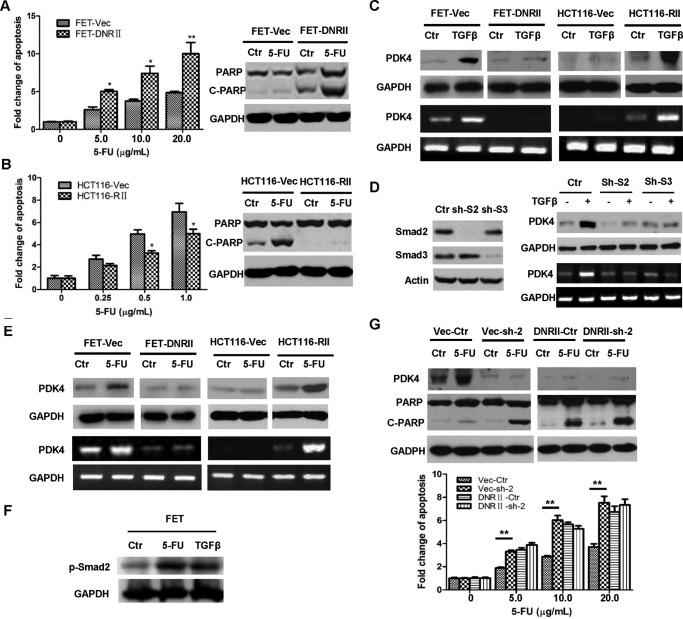
FIGURE 5.
TGFβ mediates 5-FU resistance through regulation of PDK4 expression. A, FET-Vec and FET-DNRII cells were treated with different concentrations of 5-FU for 72 h. DNA fragmentation assays were performed (left panel). Western blotting analysis of cleaved PARP (C-PARP) was performed after 2.5 μg/ml 5-FU treatment for 72 h (right panel). Ctrl, control. B, HCT116-Vec and HCT116-RII cells were treated with different concentrations of 5-FU for 72 h. DNA fragmentation assays were performed (left panel). Western blotting analysis of cleaved PARP was performed after 2.5 μg/ml 5-FU treatment for 72 h (right panel). C, FET-Vec, FET-DNRII, HCT116-Vec, and HCT116-RII cells were treated with 4.0 ng/ml TGFβ for 48 h. Western blotting (top panel) and RT-PCR (bottom panel) analyses were performed to detect PDK4 expression. D, expression of Smad2 or Smad3 was knocked down individually in FET cells (left panel). Cells were treated with 4.0 ng/ml TGFβ for 48 h. Western blotting (top right panel) and RT-PCR (bottom right panel) analyses were performed to detect PDK4 expression (right panel). E, FET-Vec, FET-DNRII, HCT116-Vec, and HCT116-RII cells were treated with 5.0 μg/ml 5-FU for 72 h. Western blotting (top panels) and RT-PCR (bottom panels) analyses were performed to detect PDK4 expression. F, FET cells were treated with 5.0 μg/ml of 5-FU or 4.0 ng/ml of TGFβ for 1 h. Western blotting analysis was performed to examine p-Smad2. G, a scrambled shRNA or PDK4 shRNA (sh-2) was infected into FET-Vec and FET-DNRII cells. The cells were treated with 2.5 μg/ml 5-FU (top panel) or different concentration of 5-FU (bottom panels) for 72 h. Western blotting analysis of PDK4 and cleaved PARP (top panels) and DNA fragmentation assays (bottom panels) were performed. The data are presented as the mean ± S.D. of triplicate experiments. *, p < 0.05; **, p < 0.01.