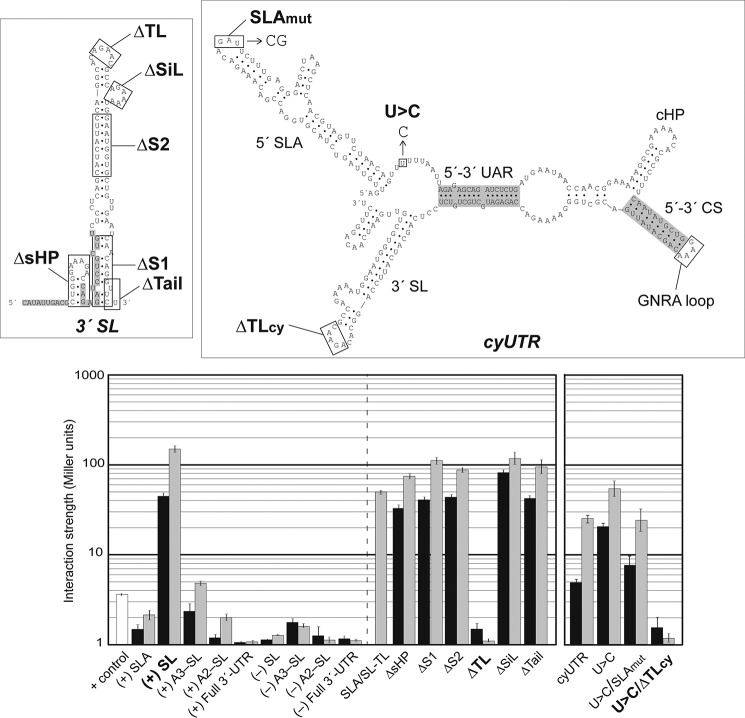
FIGURE 2.
DENV RdRp recognizes the top loop of 3′-SL. Linear 3′-SL and a series of its deletion mutants are indicated in the secondary structure with rectangles (upper left panel), whereas the model of the cyclized Y3H RNA construct containing DENV 5′- and 3′-terminal regions (cyUTR) used in this study is in the upper right panel. The long viral open reading frame was replaced by a GNRA loop that linked 5′- and 3′-UTR together in the cyUTR construct. SL mutant ΔsHP completely removed the boxed hairpin; ΔS1 removed the lower SL stem; ΔS2 removed the upper SL stem; ΔTL removed the indicated top loop nucleotides; ΔSiL removed the side loop; and ΔTail replaced the final 5′ nucleotides with CAAAA. In cyUTR, SLAmut replaced the wild-type GAU with CG, and U>C refers to the substitution of a U with a single C. Structural RNA elements are labeled. β-Galactosidase expression assays showing the interaction strengths with the full-length NS5 (black-filled bar) or RdRp (gray) are measured in Miller units. The lower left panel shows RdRp interactions with a variety of linear DENV RNAs with native sequences (left half) and mutations (right half). Results with a cyclized form of UTR are separated into the lower right panel. Error bars indicate S.E.