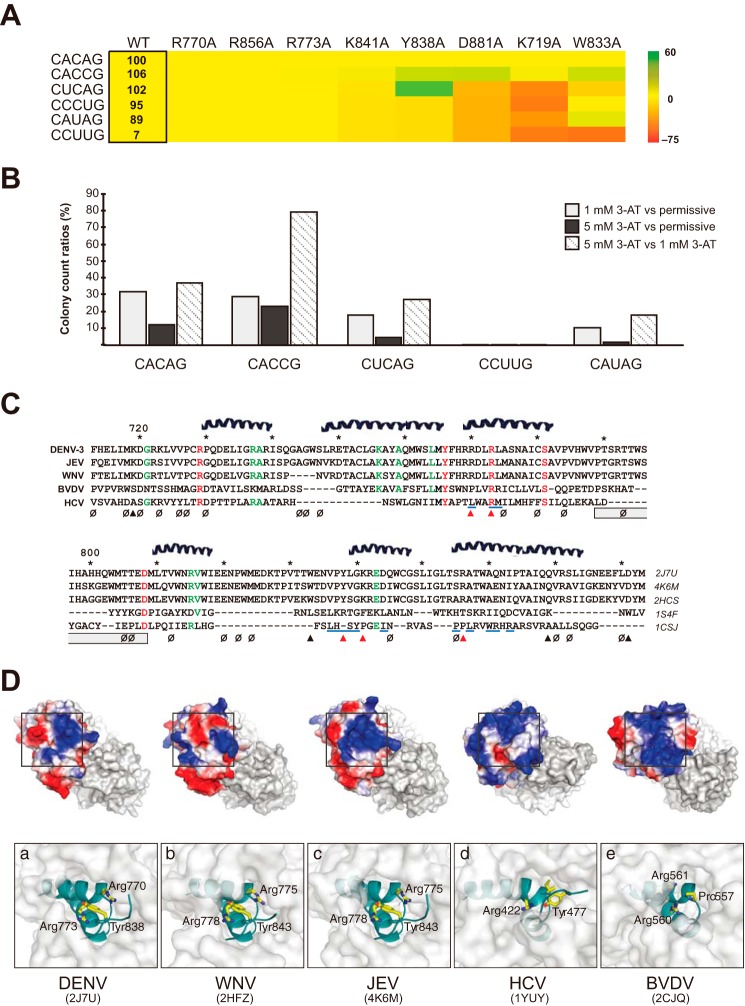
FIGURE 6.
Exploring the conservation of the RdRp-RNA interface within and beyond DENV. A, heat map matrix of interaction strengths among RdRp and 3′-SL variants. Relative interaction strengths of 3′-SL variants with the DENV RdRp are shown in a box as a percentage of the wild-type (WT) RdRp-CACAG pair. Colors, indicating a 3′-SL-TL variant's sensitivity to protein alterations, were arrived at as follows: % changes in interaction strengths between mutant RdRp and 3′-SL variant combinations versus those of WT RdRp and 3′-SL variants are calculated. These percentage changes are then compared (via simple subtraction) to the relevant RdRp-CACAG interaction strengths. Numerous RNA variants, including some residing on the 5′-SLA, are not included on the table, as their interactions with RdRp were particularly weak. B, analysis of 3′-SL-TL's tolerance for mutations in Arg-773. A library of Arg-773 RdRp mutants (R773X) was examined via Y3H with five 3′-SL-TL variants under one permissive and two selective conditions. C, DALI structural alignments of the thumb domains from five available Flaviviridae RdRp structures. Residues with perfect conservation are colored in red, and ones that align in four sequences are green. The null symbols (ø) indicate mutations that had no effect in 3′-SL-binding in our Y3H random mutagenesis. Filled triangles point to residues that were chosen for site-directed mutagenesis, and red triangles indicate crucial residues in 3′-SL-TL-binding. HCV residues making contacts with thumb 2 inhibitors in previous studies are highlighted by blue lines. The gray rectangle shows the region that was deleted in an HCV structural study (45). D, superposition of five Flaviviridae RdRp structures reveals similarity in the SL-binding site of DENV RdRp comprising an Arg-770–Tyr-838–Arg-773 stacking platform. In all cases, two non-contiguous helices combine to form a positively charged pocket (blue) centering on a perfectly conserved Arg-773 in DENV.