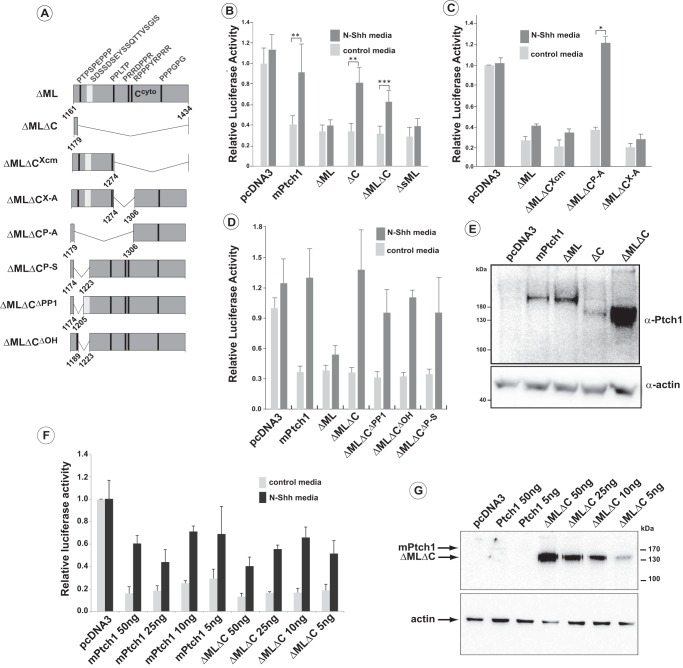
FIGURE 4.
Activities of Ptch1 deletion mutants in the regulation of Hh-signaling. A, schematic of ΔML mPtch1 mutants. All mutants harbor deletion of a.a. 614 and 709 (ΔML) in conjunction with deletion of the indicated amino acids in the C-terminal domain. B and C, repression of canonical Hh signaling and response to Hh ligand of ΔMLΔC mutants. Deletions in the C terminus responsible for restoration of the Shh signaling response showed that loss of the sequence between 1179–1306 (ΔMLΔCP-A) reverses the constitutive repression of Smo by the ΔML mutant. Sequences down stream of 1274 (ΔMLΔCX-A and ΔMLΔCXcm) had no effect on the constitutive repression activity of the ΔML mutant. Data are displayed as mean ± S.E. (n = 5 for B and 2 for C). Independent experiments were done with biological duplicates. *, p < 0.05; **, p < 0.01; ***, p < 0.001. D, further refinement of the C-terminal deletions revealed that loss of a.a. 1179–1220 (ΔMLΔCP-S), which includes a polyproline motif (PP1) and a polyhydroxyl-rich region, or the loss of either motif individually (ΔMLΔCPP1, ΔMLΔCOH) restored the Shh signaling response of ΔML. Data are displayed as mean ± S.E. (n = 4). Independent experiments were done with biological duplicates. **, p < 0.01; ***, p < 0.001. E, Western blots were run using lysates taken from the same well as that used for the luciferase reporter assay. F, luciferase activity following titration of plasmids expressing either the full-length mPtch1 or the ΔMLΔC mutant. Data are displayed as mean ± S.E. (n = 3). G, Western blotting analysis showing the relative levels of Ptch1 expression in one set of lysates used for the luciferase experiments in F.