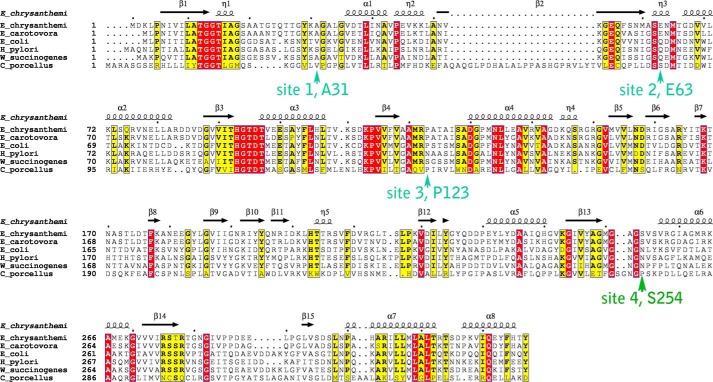
FIGURE 1.
Sequence alignment of select l-asparaginases. Strictly and mostly conserved residues are highlighted in red and yellow, respectively. Arrows indicate the four sites selected for mutagenesis. The secondary structure shown above the primary sequence is based on the atomic structure of E. chrysanthemi (PDB ID 1O7J). The E. chrysanthemi, Erwinia carotovora and E. coli enzymes contain an N-terminal signal peptide, which is not included in the alignment. The UniProt entries used for this alignment are: E. chrysanthemi, P06608; E. carotovora, Q6Q4F4; E. coli, P00805; W. succinogenes, P50286; Helicobacter pylori, O25424; Cavia porcellus (guinea pig), H0W0T5. For the guinea pig enzyme, only residues in its N-terminal l-asparaginase domain are shown.