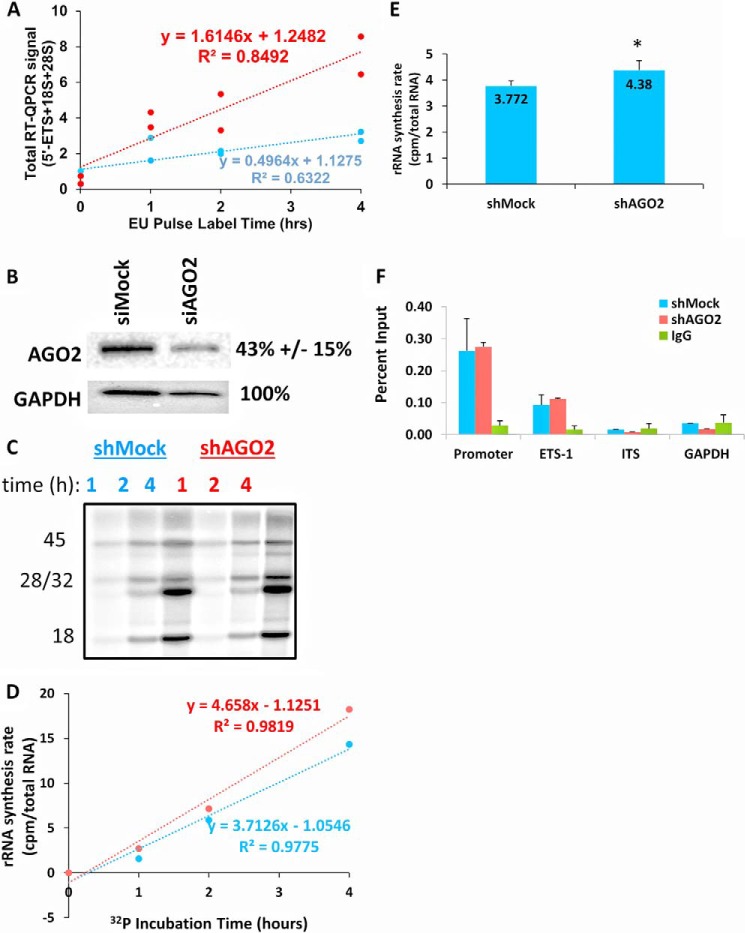
FIGURE 5.
AGO2 knockdown increases the apparent synthesis rate of rRNA. A, K562 cells were metabolically labeled with EU in both siMock- and siAGO2-treated cells for the indicated length of time (see “Experimental Procedures”). RT-QPCR was used to quantify total rRNA, normalized to GAPDH, at each time point. The time points were fitted to a linear regression to determine the rate of incorporation of EU in each treatment (1.61 versus 0.496, p = 0.00424, two-way analysis of covariance). B, a Western blot indicating the ability to knockdown AGO2 expression using the Silencer® Select siRNA (“Experimental Procedure”). A representative blot is shown, and the percentage of AGO2 knockdown is indicated to the right of the blot. In each case, AGO2 levels were normalized to GAPDH (n = 4, Student's t test). C, shMock and shAGO2 HEK293T cells were metabolically labeled with 32P for the indicated lengths of time, and total RNA was harvested, gel-electrophoresed, and imaged in a Typhoon scanner (see “Experimental Procedures”). The gel was also stained with SYBR Gold to normalize for total RNA levels. The identity of each radioactive RNA band was indicated based on its size and is indicated to the left of the gel. D, the total counts of the 45S, 32S, 28S, and 18S radioactive bands were summed, normalized to total RNA (image J software, NIH), and plotted as function of incubation time. The best fit line was determined by Microsoft Excel 2013, and the slope, y-intercept, and χ2 of the line are shown (n = three biological replicates, and three time points per replicate; *, p < 0.05, Student's t-test). E, a summary of the difference in total 32P incorporation between shMock- and shAGO2-treated HEK293T cells after 4 h of labeling (n = 5). F, A UBTF ChIP-QPCR (n = 3) was performed in both shMock and shAGO2 cells. The amplicons indicate the region of the rRNA gene that was amplified: promoter = −57, ETS1 = 851, IGS = 37997. The primer sequences were all previously reported (11).