Fig. 1.
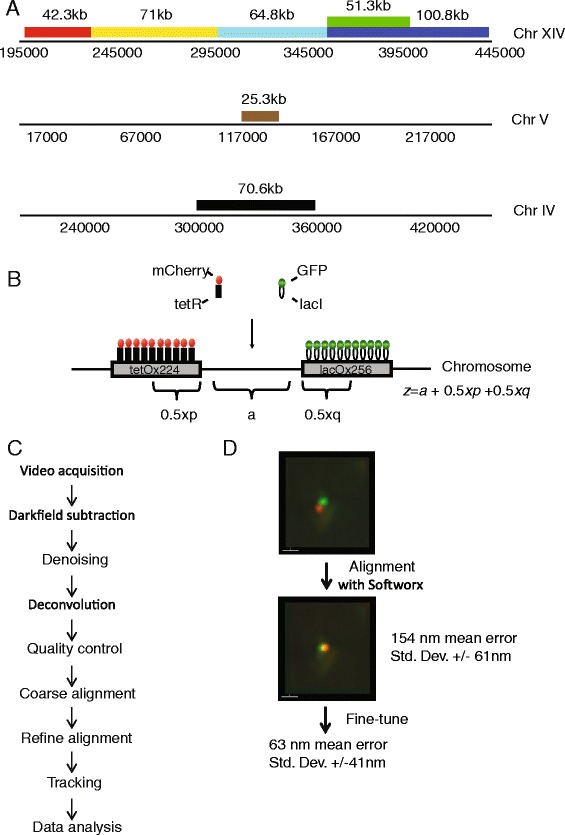
Establishment of a work-flow for live cell 4D imaging. a Seven sample strains were generated which introduced lac ×256 and tet ×224 operator arrays flanking regions on Chrs XIV, IV and V. b Operator arrays were detected using fluorescent tagged repressor proteins as indicated. A naming convention was adopted that includes the endogenous genomic distance (a) as well as half the distance of each operator array (0.5x p, 0.5x q). Not shown: An additional strain was generated with a single tetO ×224 array on Chr XIV which expressed both tetR-GFP and tetR-mCherry to produce colocalising green and red spots. This strain was used for channel alignment and mean measurement error estimation purposes. c Summary of the work flow for image processing. d Two stage channel alignment was found to improve standard error from 154 nm to 63 nm
