Fig. 6.
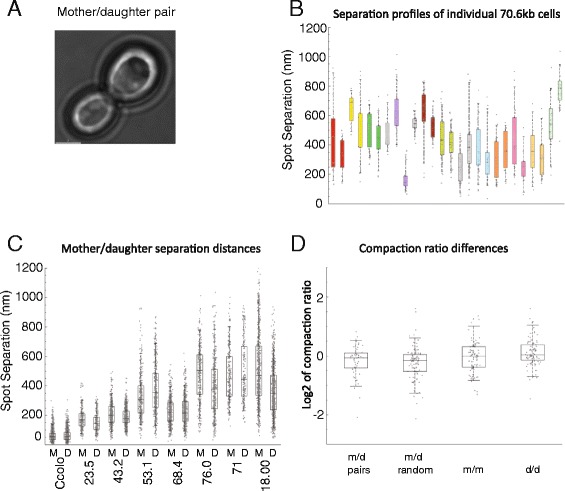
Chromatin conformation is not inherited. Mother and daughter cells can readily be identified in Saccharomyces cerevisiae cultures as physically attached pairs (a). Each mother-daughter pair shares the same parent providing an opportunity to assess whether chromatin configuration is conserved through cell division. Movies obtained in individual cells from the 70 kb strain are plotted with mother-daughter pairs in the same colour in (b), in each case with data from the mother to the left. Unpaired cells are in grey. In some cases it is clear that mothers and daughters have differing spot distances. The distance box plots for all mothers and daughters of each strain are plotted in (c). In order to compare changes in distance between a large sample of related and unrelated mother and daughter cell pairs, changes in compaction were calculated using movies from each genomic separation (d). Compaction is calculated by dividing genomic separation by the measured distance between operators. The change in compaction between different populations was then calculated as a ratio indicated on the y-axis as ‘Log2 of compaction ratio’. This comparison could be made between related m/d pairs and unrelated m/d pairs selected at random. In both cases, the distributions of the differences in compaction are similar and not statistically significant (Mann-Whitney p-value = 0.181). This indicates that the separation of the reporter loci studied here is not inherited. Little change in separation is observed when the comparison is made between unrelated mother/mother and daughter/daughter pairs. This suggests that the difference in the volume of mother and daughter cell nuclei has little effect on the separation of these operator tagged loci
