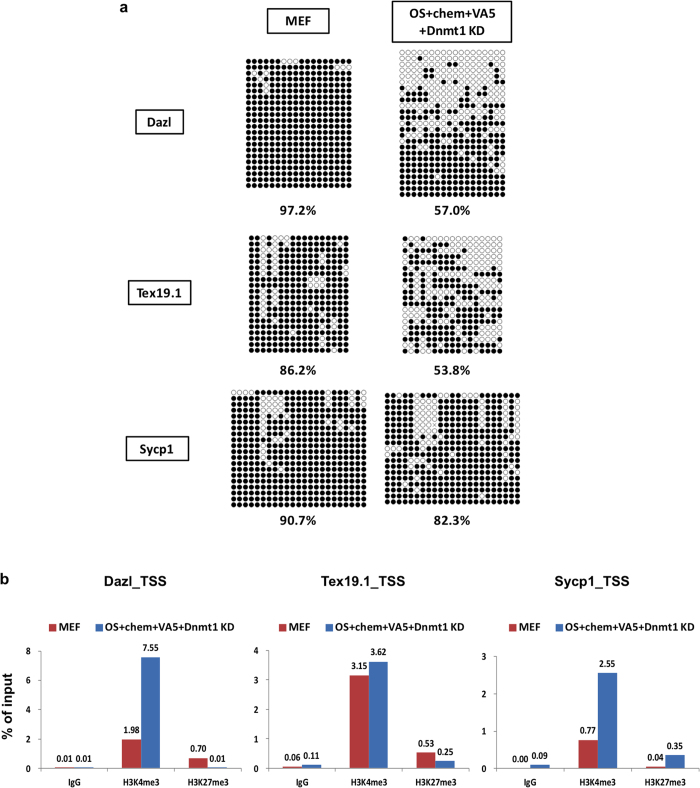
Figure 6. Changes in epigenetic modifications in MEFs following OS + chem + VA5 + Dnmt1-KD treatment.
(a) The DNA methylation status in the Dazl, Tex19.1, and Sycp1 promoter regions in MEFs 4 days after the OS + chem + VA5 + Dnmt1-KD treatment or in control MEFs. DNA methylation status was determined by bisulphite sequencing. The filled and open circles indicate methylated- and un-methylated CpGs, respectively. The data shown were combined from two independent experiments. The percentage of methylated-CpGs is shown. (b) ChIP analysis of the promoter regions of Dazl, Tex19.1, and Sycp1 for H3K4me3 and H3K27me3 in MEFs 4 days after the OS + chem + VA5 + Dnmt1-KD treatment or in control MEFs. ChIP using normal IgG was used as negative control. Levels of H3K4me3 or H3K27me3 were determined by real-time PCR, and the percentages of values relative to those for input chromatin are shown. Representative data from two independent experiments are shown. See also Supplementary Fig. S9.