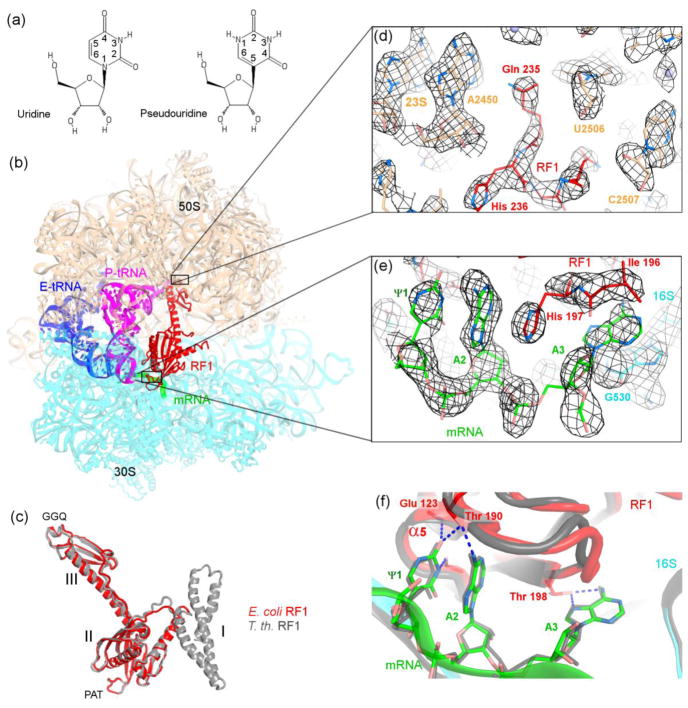
Figure 1.
Structural basis for recognition of the ΨAA codon by release factor 1. (a) Chemical structures of pseudouridine and uridine. (b) Crystal structure of the 70S•ΨAA•RF1 termination complex (this work). The 50S subunit is shown in wheat and 30S subunit in cyan. (c) Similar conformations of E. coli (this work) and T. thermophilus release factors in 70S termination complexes. (d–e) Unbiased feature-enhanced density map [37] shows well-resolved features of RF1 in the peptidyl-transferase center in (d) and ΨAA and RF1 in the decoding center in (e). (mRNA and RF1 were not included in the simulated-annealing-refined 70S model used for map calculation.) (f) Comparison of the conformations of the ΨAA (this work) and UAA (gray) codons in the 70S•RF1 termination complexes. In panels (c) and (f), the structure of T. thermophilus 70S•RF1 complex (PDB ID: 4V63, [11]) is shown in gray.