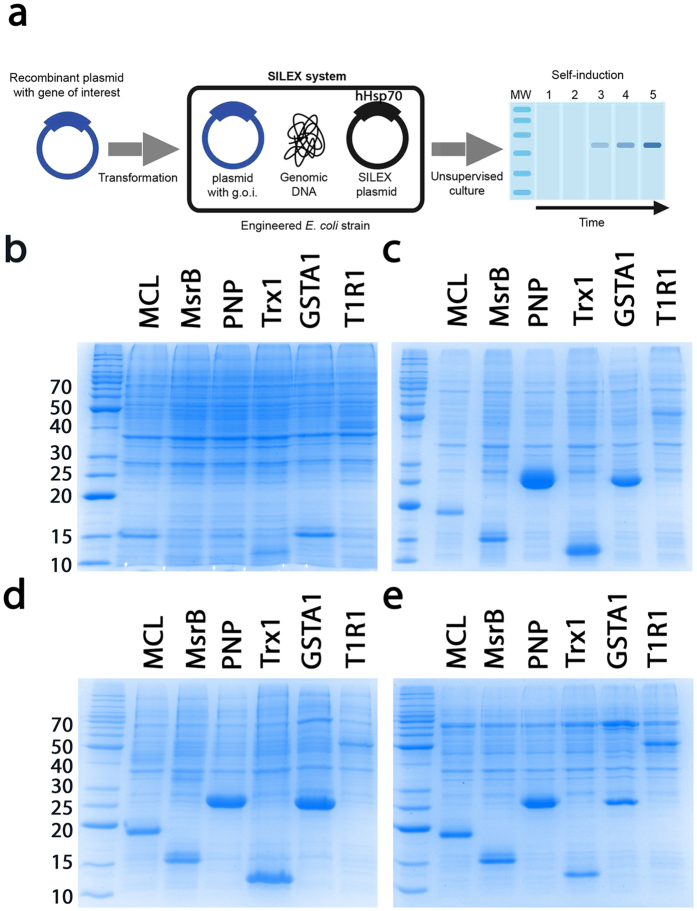
Figure 3. Principle of the SILEX system and application to 6 different recombinant proteins compared to the other primary expression system.
(a) The use of SILEX can be divided in three steps, Step 1: As with other methods, a plasmid containing the coding sequence of the protein of interest (blue) is introduced (step 2) into host cells engineered from an E. coli strain containing a SILEX plasmid (black). Step 3: SDS-PAGE illustrating autoinduction in the SILEX system without the need to monitor the cell density or add a chemical inducer. Protein expression was monitor on SDS-PAGE after (b) BL21(DE3) growth without induction, (c) BL21(DE3) growth with IPTG induction, (d) BL21(DE3) growth in the ZYM auto-inducible medium, and (e) SILEX growth without any inducer addition. For each of the 6 proteins (Richardella dulcifica miraculin (MCL), Xanthomonas campestris methionine sulfoxide reductase B (MsrB), E. coli purine nucleoside phosphorylase (PNP), E. coli thioredoxin 1 (Trx1), H. sapiens glutathione transferase A1 (GSTA1) and H. sapiens taste receptor type 1 member 1 (T1R1), a cell aliquot was subjected to SDS-PAGE for expression analysis. The molecular weight markers are indicated on the left of the gels in kDa.