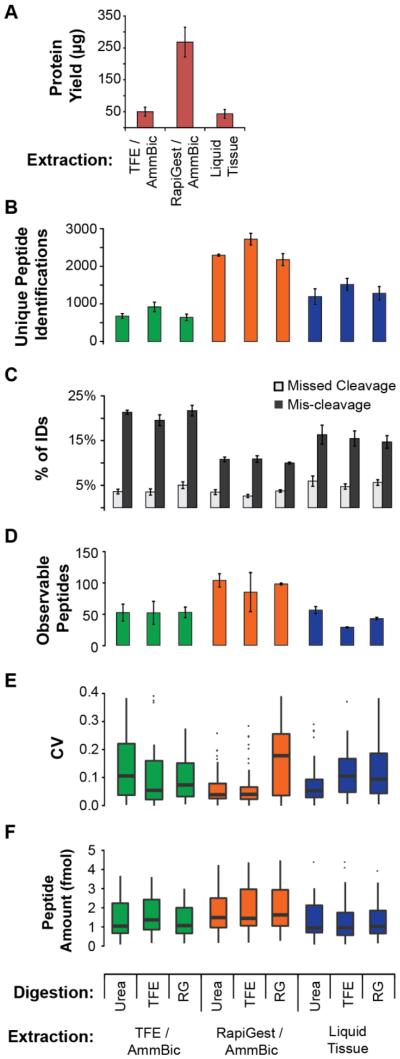
Figure 2. Evaluation of processes for quantitative proteomics on FFPE.
Results are from the analysis of an FFPE, ER+/HER2+ breast cancer. Each sample consists of three 10 μm sections. All error bars reflect the standard deviation of three analytical process replicates (i.e. de-paraffinization, protein extraction, digestion, enrichment, analysis performed on three separate days). A. Protein yield measured by micro-BCA assay. B. Unique peptide identifications obtained from shotgun LC-MS/MS results. Equal amount of protein (2 μg) was loaded on-column for each process. C. Trypsin digestion efficiency and fidelity measured by the percentage of identifications containing missed cleavages (internal K or R) or mis-cleavages (non-tryptic cut site). D. LC-MRM results showing the number of observable peptides (>LOD in 2 out of 3 replicates). Heavy peptide standards were added to1 μg of the digested lysates at 10 fmol/μg protein and analyzed in process triplicate. E. LC-MRM results showing the distribution of CVs for observable peptides. F. Distribution of peptide amounts measured by LC-MRM. Box plots show the median (bar), inner quartiles (box), 5-95th percentiles (line), and outliers (points).