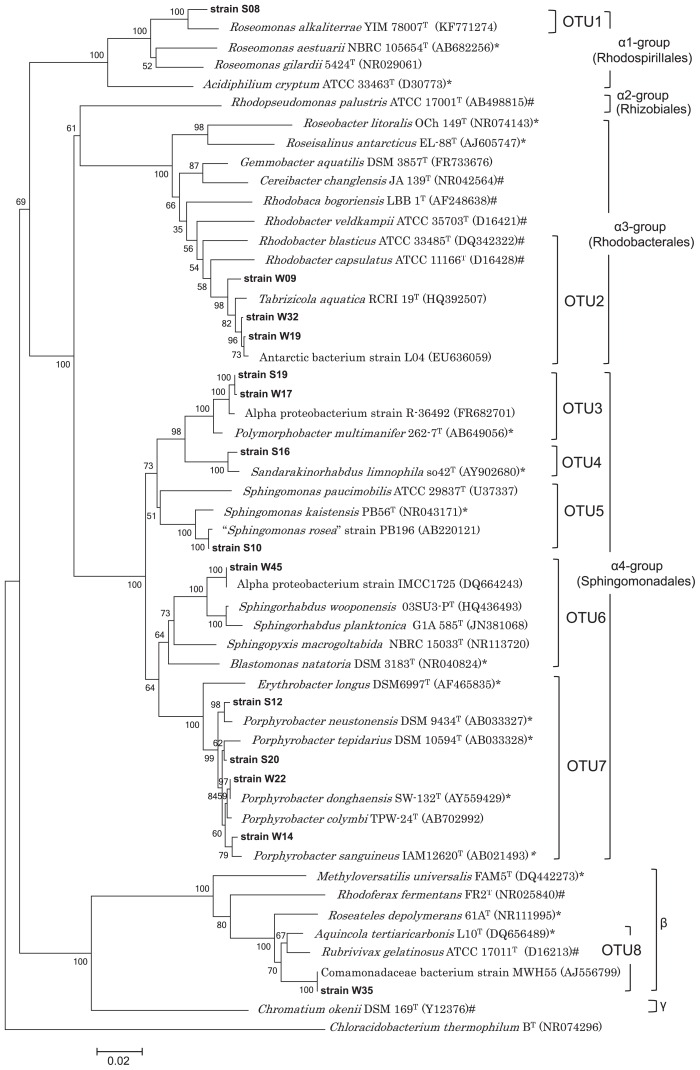
Fig. 2.
Neighbor-joining phylogenetic tree based on 16S rRNA gene sequences (1,409 positions). The 16S rRNA gene sequences of strains isolated in this study are indicated in bold by the “strain W” and “strain S” prefix in the case of winter and summer isolates, respectively. Sequences from the database are represented with their respective accession numbers behind species and strain names in parentheses. Among these, bacteria known to be AAP bacteria were marked with an asterisk (*) beside the accession numbers. Bacteria known to be anaerobic anoxygenic photosynthetic bacteria (purple photosynthetic bacteria) were marked with a hash (#) beside the accession numbers. α1-group, α2-group, α3-group, and α4-group shown in the right of the tree indicate groups (orders) of the class Alphaproteobacteria. β and γ indicate the classes Betaproteobacteria and Gammaproteobacteria, respectively. The scale bar represents the number of substitutions in each site. Bootstrap values at branch points are expressed as percentages of 500 resamplings. Chloracidobacterium thermophilum strain BT (NR074296) was used as an outgroup to root the tree.