Figure 2. Genomic sequence features of ProtoRAG.
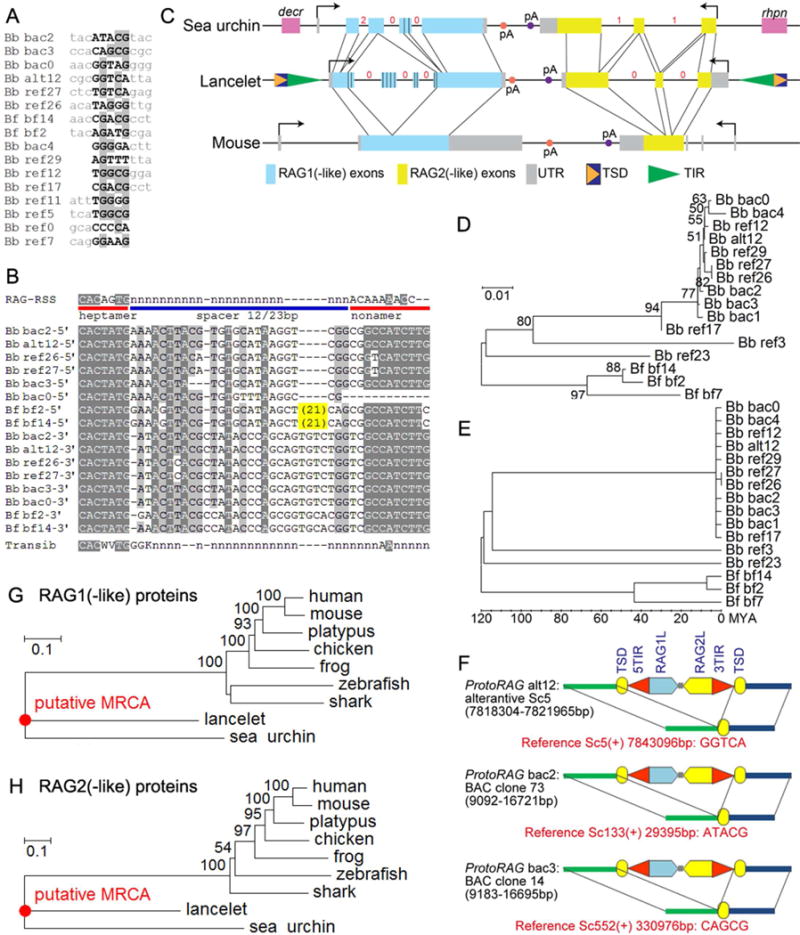
(A) Alignment of ProtoRAG TSDs and flanking sequences from the lancelet genome.
(B) Alignment of ProtoRAG TIR sequences with the consensus RSS and Transib TIR. IUPAC codes used in the alignment: N=A, C, G or T; K=G or T; W=A, T; V=A, C or G. Lower case n indicates an undetermined nucleotide. Shading indicates sequence conservation, with darker gray indicating a higher degree of conservation. Bb: B. belcheri; Bf: B. floridae. ProtoRAG copy identification numbers correspond to those listed in Table S1.
(C) Genomic organization of the lancelet ProtoRAG copy on BAC plasmid clone 73, sea urchin RAG1/2-like gene locus, and mouse RAG locus. Corresponding coding regions are indicated by thin lines. The phases of introns in coding regions are shown by red numbers. Repetitive regions in lancelet RAG1-like and sea urchin RAG1-like are marked by vertical bars. Terminal exons of flanking genes (decr, dienoyl-CoA reductase; rhpn, rhophilin) for the sea urchin RAG1/2-like locus are shown as purple boxes.
(D) Neighbor-Joining trees of lancelet ProtoRAG copies assembled using Mega v5.2 (see Supplementary Methods).
(E) Molecular dating analysis of lancelet ProtoRAG copies. This linearized tree with clock calibration was calculated using Mega v5.2. The root, or the divergence between B. belcheri and B. floridae, was set to 120 million years ago.
(F) Three unfixed (polymorphic) ProtoRAG transposition events identified in B. belcheri genomes. Red text provides the coordinates of target sites and the sequence of TSDs on the reference genome. See Table S1 and Data S1 for further details.
(G, H) Neighbor-joining trees of RAG1 (G) and RAG2 (H) protein homologs. MCRA, most common recent ancestor.
See also Data S1 and Table S1.
