Figure 4. TIR-dependent transposon excision mediated by bbRAG1L/2L ex vivo.
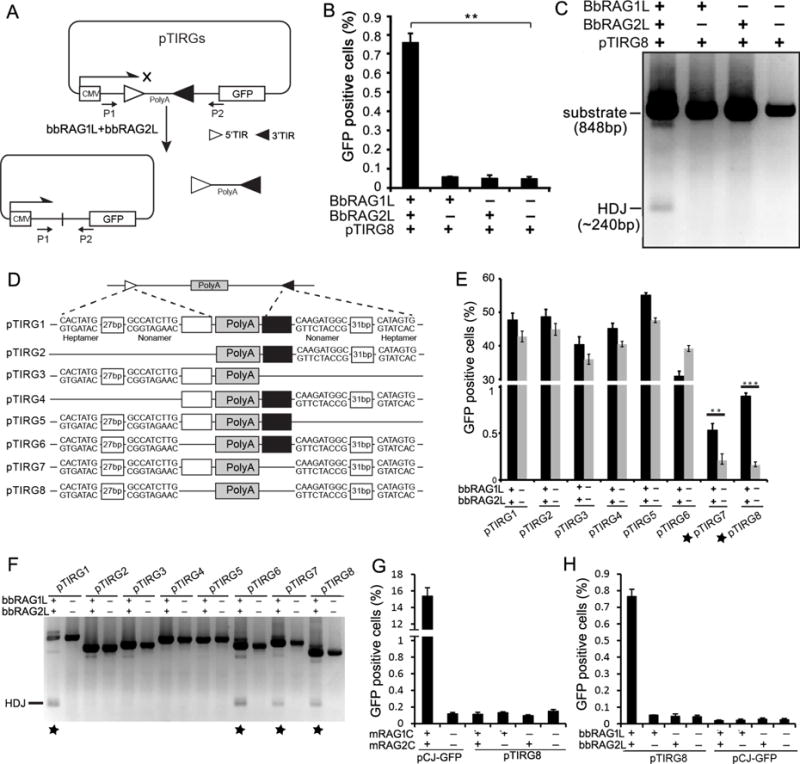
(A) Diagram of the cell-based fluorescent reporter and PCR assay for DNA excision and recombination. Filled and unfilled triangles, 5′- and 3′-TIR sequences of ProtoRAG, respectively; P1/P2, PCR primers; CMV: cytomegalovirus promoter; PolyA: polyadenylation signal sequence.
(B) Quantification of GFP positive cells by flow cytometry after transfection of 293T cells with pTIRG8 (containing the minimal ProtoRAG TIRs) with bbRAG1L and bbRAG2L expression vectors, as indicated.
(C) PCR detection of HDJs from transfections of pTIRG8 as in (B).
(D) Diagram of truncated TIR-containing substrates. Unfilled and filled boxes indicate the remainder of the 5′- and 3′-TIRs, respectively.
(E) Quantification of GFP positive cells by flow cytometry after transfection of 293T cells with truncated TIR-containing substrates.
(F) PCR detection of HDJs for truncated TIR-containing substrates.
(G) Quantitation of GFP positive cells by flow cytometry after transfection of 293T cells with mouse RAG1 core and RAG2 core expression vectors with pTIRG8 (containing the minimal ProtoRAG TIRs) or pCJ-GFP (containing RSSs), as indicated.
(H) Quantitation of GFP positive cells by flow cytometry after transfection of 293T cells with bbRAG1L and bbRAG2L expression vectors with pTIRG8 or pCJ-GFP (which contains a 12/23 RSS pair instead of the TIRs of pTIRG8), as indicated.
See also Figure S3 and S4.
