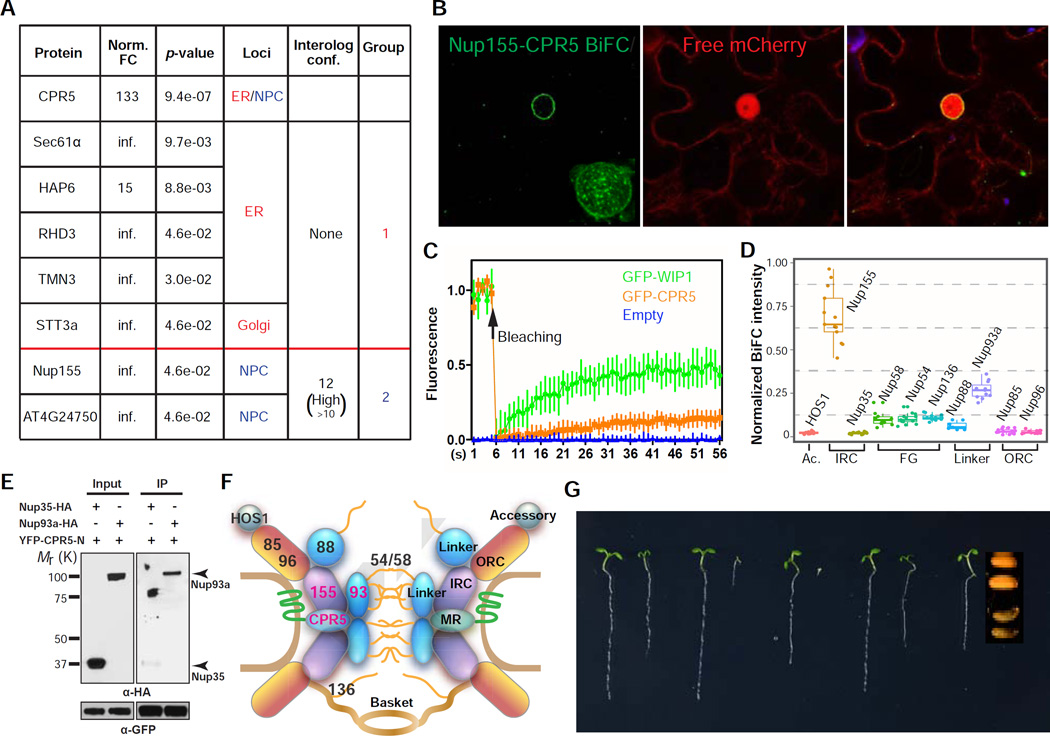
Figure 2. CPR5 Physically and Genetically Interacts with Nucleoporins as a Component of the NPC.
(A) CPR5 interactors identified by protein complex purification followed by LC-MS/MS. FC, fold change of spectrum counts in YFP-CPR5 vs GFP sample. Infinite (inf.) indicates that peptide was not detected in GFP samples. Interolog conf., confidence of predicted interaction between proteins.
(B) CPR5 and Nup155 interacts in the NE. Bimolecular fluorescence complementation (BiFC) assay was performed by transiently coexpressing nYFP-CPR5 and Nup155-cYFP in N. benthamiana. The interaction pattern on the nuclear surface was reconstructed by Z-stack images (inset).
(C) Fluorescence Recovery After Photobleaching (FRAP) analysis of GFP-CPR5 in transgenic Arabidopsis. A mobile NE protein GFP-WIP1 served as a control. Data are presented as mean ± SDM (n = 5 experimental replications).
(D) Interaction mapping of CPR5 with nucleoporins. BiFC was performed by transiently coexpressing nYFP-CPR5 with Nup-cYFP in N. benthamiana. The BiFC intensity was normalized using averaged expression levels of corresponding Nup-YFP measured in separate experiments. Ac, accessory nucleoporin; IRC, inner ring complex; FG, Phe-Gly repeat-containing nucleoporin; ORC, outer ring complex; Linker, linker nucleoporin.
(E) CPR5 interacts with the IRC-associated linker nucleoporin Nup93a. In vitro pulldown assay was performed using GFP-TrapA agarose beads. YFP-CPR5-N, YFP-tagged N-terminal half of CPR5.
(F) The structural modules of the nuclear pore complex (NPC) and the proposed position of CPR5 within the NPC.
(G) Genetic interaction between cpr5 and mutants of the ORC nucleoporins. 5-day-old seedlings were shown. Since the cpr5 nup160 double mutant did not germinate, the seed morphology of the homozygote was compared to that of a heterozygote.
See also Figure S2.