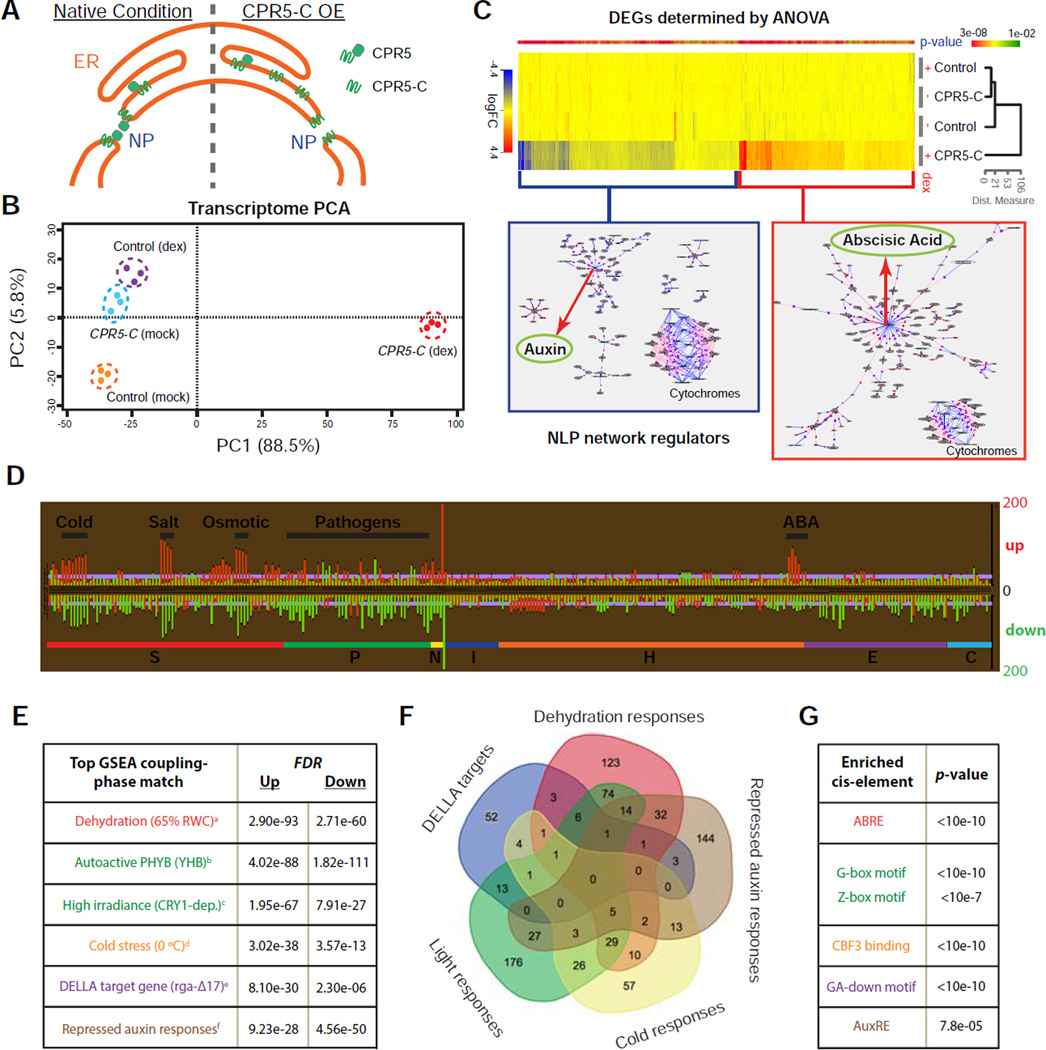
Figure 3. Transient Interference with CPR5 Function Simultaneously Activates Diverse Nuclear Signaling Pathways.
(A) A proposed cellular mechanism for transient interference of CPR5 function by overexpression of CPR5-C. NP, nuclear pore.
(B) Principal component analysis (PCA) of transcriptome changes induced by transient expression of CPR5-C.
(C) Heat map of differentially expressed genes (DEGs) that depend on CPR5-C transgene induction (p-value < 0.01). Genes with fold change (dex vs mock) higher than 2 were subject to natural language processing (NLP)-based network regulator discovery analysis. Major network regulators are highlighted with red arrows.
(D) Comparative analysis of CPR5-C-mediated CPR5 interference data with 438 published Arabidopsis microarrays. Each row represents an analysis with a specific array dataset, and datasets are sub-categorized into sections according to treatments. Section code: S, stress; P, pathogen; N, nutrient; I, inhibitor; H, hormone; E, elicitor; C, chemical. The length of each bar represents the number of overlapping DEGs (up- or down-regulated) between the CPR5-C data and a specific treatment.
(E) Gene Set Enrichment Analysis (GSEA). CPR5-C-induced up- and down-regulated genes were used as separate inputs for GSEA and non-redundant top matches are listed. PubMed IDs for the listed datasets are: a-21050490, b-19529817, c-17478635, d-16214899, e-17933900, f-19392692.
(F) Venn diagram of the transcriptome signatures identified in (E).
(G) Cis-element enrichment analysis of CPR5-C-induced total DEGs. ABRE, ABA-responsive element; G/Z-box, light-responsive elements; CBF3, transcription factor for cold acclimation; GA-down, gibberellin down regulated d1 cluster; AuxRE, auxin response element.