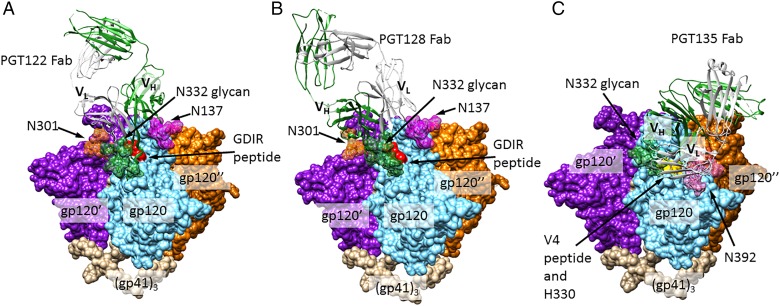
Fig. 3.
N332-dependent bnAbs. (A) Crystal structure of PGT122 Fab bound to BG505 SOSIP.664 gp140 trimer, a native-like, cleaved trimeric HIV Env construct (PDB ID: 4NCO). N301, 322 and 137 glycans are shown in forest green, orange and magenta, respectively, with all other glycans removed for clarity. Identically bound PGT122 Fabs on the other two gp120 protomers are also not shown. (B) A model of the PGT128-HIV Env interaction, created by aligning a gp120 chain from the BG505 Env structure (PDB ID: 4NCO) with the gp120 chain of a PGT128-eODmV3 gp120 co-crystal structure (PDB ID: 3TYG). N332 and 301 glycans depicted (forest green and orange, respectively) are from the PGT128 structure and N137 glycan (magenta) is from the BG505 structure. (C) A model of the PGT135-HIV Env interaction, created in a manner analogous to (B). PGT135 and the N332 and 392 glycans (forest green and pink, respectively) are from a structure of the quaternary complex PGT135 + gp120 core + CD4 + 17b Fab (PDB ID: 4JM2). All glycans outside the bnAb epitopes are omitted for clarity, as are the two Fabs binding other gp120 protomers. This figure is available in black and white in print and in color at Glycobiology online.