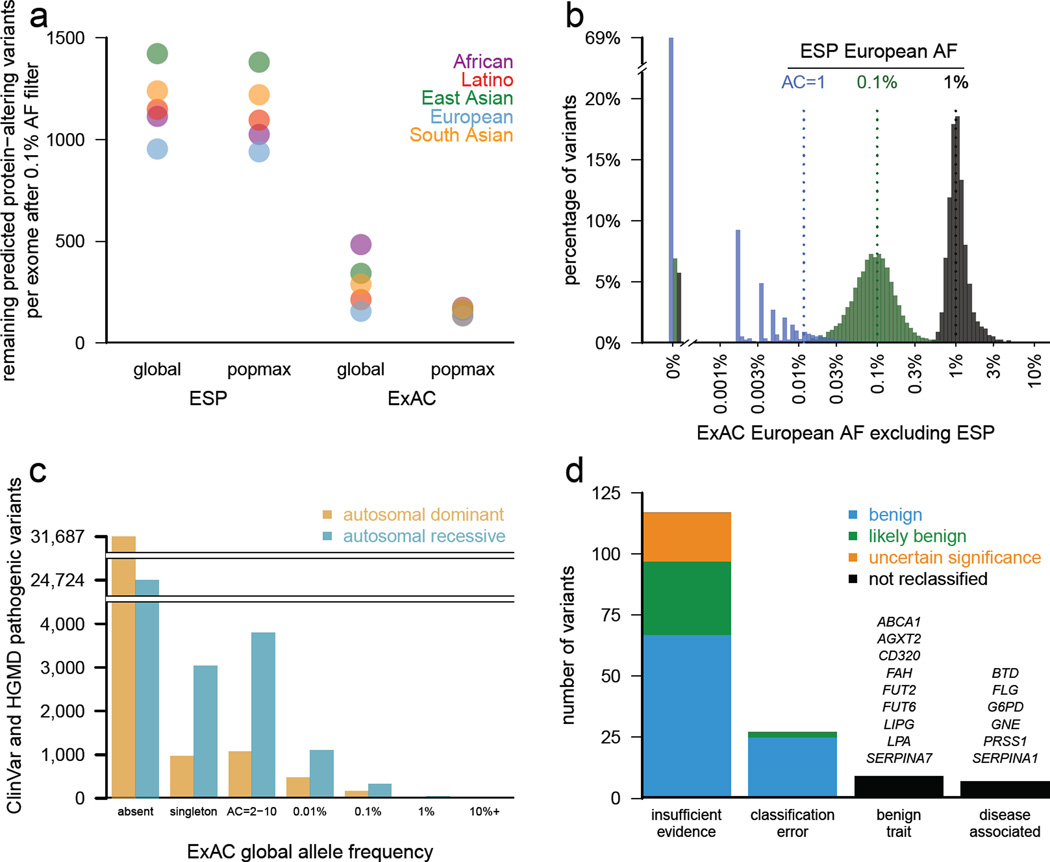
Figure 4. Filtering for Mendelian variant discovery.
a) Predicted missense and protein-truncating variants in 500 randomly chosen ExAC individuals were filtered based on allele frequency information from ESP, or from the remaining ExAC individuals. At a 0.1% allele frequency (AF) filter, ExAC provides greater power to remove candidate variants, leaving an average of 154 variants for analysis, compared to 1090 after filtering against ESP. Popmax AF also provides greater power than global AF, particularly when populations are unequally sampled. b) Estimates of allele frequency in Europeans based on ESP are more precise at higher allele frequencies. Sampling variance and ascertainment bias make AF estimates unreliable, posing problems for Mendelian variant filtration. 69% of ESP European singletons are not seen a second time in ExAC (tall bar at left), illustrating the dangers of filtering on very low allele counts. c) Allele frequency spectrum of disease-causing variants in the Human Gene Mutation Database (HGMD) and/or pathogenic or likely pathogenic variants in ClinVar for well characterized autosomal dominant and autosomal recessive disease genes28. Most are not found in ExAC; however, many of the reportedly pathogenic variants found in ExAC are at too high a frequency to be consistent with disease prevalence and penetrance. d) Literature review of variants with >1% global allele frequency or >1% Latin American or South Asian population allele frequency confirmed there is insufficient evidence for pathogenicity for the majority of these variants. Variants were reclassified by ACMG guidelines24.