Abstract
Objective
Monogenic diabetes is rare but an important diagnosis in pediatric diabetes clinics. These patients are often not identified as this relies on recognition of key clinical features by an alert clinician. Biomarkers (islet autoantibodies and C-peptide) can assist in exclusion of patients with Type 1 diabetes and allow systematic testing that does not rely on clinical recognition. Our study aimed to establish the prevalence of monogenic diabetes in UK pediatric clinics using a systematic approach of biomarker screening and targeted genetic testing.
Research design and methods
We studied 808 patients (79.5% of the eligible population) <20 years of age with diabetes attending six pediatric clinics in South West England and Tayside Scotland. Endogenous insulin production was measured using the Urinary C-peptide creatinine ratio (UCPCR). C-peptide positive patients (UCPCR ≥0.2nmol/mmol) underwent islet autoantibody (GAD and IA-2) testing, with negative cases undergoing genetic testing for all 29 identified causes of monogenic diabetes.
Results
2.5% (20/808), (95% confidence interval (CI) 1.6-3.9 %), of patients had monogenic diabetes (8 GCK, 5 HNF1A, 4 HNF4A, 1 HNF1B, 1 ABCC8, 1 INSR). The majority, 17/20, were managed without insulin treatment. A similar proportion of the population had Type 2 diabetes (3.3%, 27/808).
Conclusion
This large systematic study confirms a prevalence of 2.5% with monogenic diabetes aged <20yrs in 6 UK clinics. This figure suggests that around 50% of the estimated 875 UK pediatric patients with monogenic diabetes are still not diagnosed. This biomarker screening pathway is a practical approach that can be used to identify pediatric patients most appropriate for genetic testing.
Background
Monogenic diabetes is often not recognised in children or adolescents and misdiagnosis as Type 1 in these individuals is common (1–8). Making the correct diagnosis of monogenic diabetes is vitally important as the management of the commonest subtypes (GCK, HNF1A and HNF4A maturity onset diabetes of the young (MODY)) is markedly different from Type 1 diabetes (9,10). Molecular diagnosis improves clinical care by confirming the diagnosis, aiding prediction of the expected clinical course of the disease and guiding appropriate management and family follow up (10–12). Due to the predominance of Type 1 diabetes in children, the potential significance of a parent with diabetes or possible non-insulin dependence may be overlooked. This leads to unnecessary insulin treatment with a mean delay from diabetes diagnosis to the correct genetic diagnosis of 9.3 years (unpublished data KC, SE), (based on 1240 patients initially diagnosed with diabetes <20 years and having a genetic diagnosis of GCK, HNF1A or HNF4A MODY).
The present approach to diagnosing monogenic diabetes requires clinical recognition by an alert health care professional and subsequent genetic testing. As genetic testing for monogenic diabetes is now widely available worldwide the major barrier is clinician recognition (although costs and lack of medical insurance coverage of genetic testing can also limit who is tested). Despite the availability of guidelines advising when a diagnosis of monogenic diabetes in children should be suspected (10) genetic testing is under requested. We have shown that under diagnosis of MODY in some regions in the UK reflects reduced testing rather than inappropriate testing (13).
Biomarker tests can help identify appropriate candidates for genetic testing for monogenic diabetes, avoiding reliance on clinical recognition. These biomarkers are most useful in enabling a firm diagnosis of Type 1 diabetes to be made, obviating consideration of genetic testing. The lack of significant endogenous insulin production (stimulated serum C peptide <200pmol/l) is seen in Type 1 diabetes outside the honeymoon period. Urinary C-peptide creatinine ratio (UCPCR) provides a simple, stable, reliable, non-invasive measure, which can be tested on a sample posted from home direct to a laboratory (14,15) and has been validated against the mixed meal tolerance test (16). UCPCR ≥0.2nmol/mol indicates endogenous insulin and has been used to differentiate patients with MODY from Type 1 diabetes >5 years post diagnosis (17). Islet autoantibodies are found in the majority of Type 1 patients especially when measured close to diagnosis and are an excellent discriminator between Type 1 diabetes and MODY (18).
A large number of studies have tried to assess the prevalence of monogenic diabetes in the pediatric population (Table 1), however the majority of these studies do not use a systematic approach or are limited to single clinic populations. A further limitation is that no studies to date have investigated all the causes of monogenic diabetes (Table 1). Only three studies have systematically screened large populations: i) a US multicentre systematic study (SEARCH) identified a minimum prevalence of 1.2% with MODY (1) and a further 0.2% with neonatal diabetes (19), ii) a Norwegian nationwide study identified a minimum prevalence of monogenic diabetes in children of 1.1% (2), iii) a Polish study identified a minimum prevalence of 3.1-4.2% (7). Other smaller studies (4, 8, 20–22) report screening or assessment of single pediatric clinic populations and although islet autoantibody negativity is often used to identify children who could benefit from genetic testing the screening and testing strategies are variable with estimates of prevalence up to 2.5%. Survey/questionnaire or epidemiological data relying on physician reporting and recognition of clinical features of monogenic diabetes in pediatric populations state widely varying prevalence of 0.6-4.2% (7, 23–27). However these approaches do not involve systematic screening and therefore may be considered less accurate.
Table 1.
Approaches used to identify monogenic diabetes in pediatric populations
| Type of study | Country | Area | Initial cohort (n) | Cohort characteristics | Testing strategy (subgroup tested) | Genes tested | Prevalence in genetically tested | Minimal prevalence of monogenic diabetes | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Systematic studies ordered by number in study | |||||||||
| Multi-centre population based | USA | 6 centres: California, Ohio, Hawaii, South Carolina, Washington | 5963 | 1) Diagnosed <20yrs 2) Diagnosed<6mths |
1) AB-ve (x2), fasting c-peptide ≥0.8ng/ml (n=586) 2) Diagnosed <6mths (n=7) |
1) HNF1A, HNF4A, GCK, 2) KCNJ11, INS, ABCC8 |
1) 8.4% (47/586) 2) 71.4% (5/7) |
1.2% 0.2% (Total 1.4%) |
Pihoker 2013 Shankar2013 |
| Nationwide population based | Norway | Nationwide | 2756 | Newly diagnosed aged 0-14 yrs | 1) AB-ve (x2) and affected parent (n=46) 2) AB-ve, HbA1c <7.5% (58mmol/mol) and not on insulin (n=10) 3) diagnosed <12 mths (n=24) |
1) HNF1A, HNF4A, MIDD 2)GCK, 3)KCNJ11, ABCC8, INS |
1) 13.0% (6/46) 2) 30.0% (3/10) 3) 16.6% (4/24) |
1.1% | Irgens 2013 |
| Epidemiological data / nationwide genetic test results | Poland | 3 centres: Lodz, Katowice, Gdansk | 2568 | Aged 0-18 yrs | 1) AB-ve, affected parent, non insulin dependent 2) HbA1c<7.5% (58mmol/mol) 3) Diagnosed <6mths 4) Syndromic diabetes |
1)HNF1A, HNF4A, HNF1B, 2)GCK 3)KCNJ11, ABCC8, INS, 4)WFS, Alstrom |
32.1% (100/311) | 3.1-4.2% | Fendler 2012 |
| Single pediatric clinic population | USA | New York | 939 | Clinical diagnosis T1D Aged 6mths-20yrs |
AB-ve (x3) plus either HbA1c≤7% (53mmol/mol) and ≤0.5u insulin /kg/day / > 1yr post diagnosis c-peptide+ or 3 gen. FH (n=58) | GCK HNF1A |
8.6% (5/58) | 0.5%* | Gandica 2015 |
| Pediatric clinics in single city | Australia | Sydney | 497 | 1) Clinical diagnosis T1D 2) Diagnosed 6mths – 16 yrs |
AB-ve (x4- on 2 occasions (n=19) | 1) HNF1A, HNF4A, 2) INS, KCNJ11 |
5% (1/19) | 1.2%* | Hameed 2010 |
| Single pediatric clinic population | Spain | Madrid | 252 | 1) Clinical diagnosis T1D 2) Diagnosed 6mths - 17yrs of age |
AB-ve (x5) (n=25) | 1)HNF1A, HNF4A, 2)KCNJ11, INS |
8.0% (2/25) | 0.8%* | Rubio-Cabezas 2009 |
| Pediatric clinic: Case Histories | New Zealand | South Island | 160 | Pediatric diabetes <18yrs | AB-ve ( x2?) (n=4) | GCK, HNF1B, HNF1A | 2.5% (4/160) | 2.5% | Wheeler 2013 |
| Nationwide | Japan | Centres throughout Japan | N/K | Aged 6mths -20yrs | 1) AB-ve (x 2), BMI<25, dominant family history or 2) renal cysts (n=80) |
1) HNF1A, GCK, HNF4A, MIDD, 2) HNF1B |
47.5% (38/80) | - | Yorifuji 2012 |
| Single pediatric clinic population | USA | Colorado | N/K | Diabetes <25 yrs | c-peptide ≥0.1ng/ml, AB-ve (x3) (n=97) |
HNF1A, HNF4A, GCK, PDX1, HNF1B | 22.7% (22/97) | N/K | Chambers 2015 |
| Non systematic studies relying on clinical recognition and clinical testing | |||||||||
| Type of study | Country | Area | Initial cohort of subject with diabetes and the population taken from (n) | Cohort characteristics | How monogenic diabetes was defined | Number with monogenic diagnosis (% all diabetes) | Prevalence per 100,000 population | Reference | |
| Postal questionnaire survey | UK | Nationwide | 15,255 (59M pop ) | Diabetes <16 yrs ′non T1′ | Confirmed by genetic test | 20 (0.13%) | 0.17 | Ehtisham 2004 | |
| Questionnaire and telephone survey | Germany | State of Baden-Württenberg | 2640 (2.6M) pop | 0-20yrs | Clinician diagnosis (45% genetically confirmed) | 58 (2.1%) | 2.3 | Neu 2009 | |
| Assessment of Childhood Diabetes registry | Germany | Saxony (34 paed clinics) | 865 new cases Prevalence cases not stated (4.8M pop) |
Newly diagnosed aged 0-15yrs | Confirmed by genetic test | 21 (2.4%) prevalence in incident cases | Cannot be calculated | Galler 2009 | |
| Surveillance questionnaire (Physician reporting) | Canada | National | Not stated (35M pop Canada) | Newly diagnosed non-type 1 diabetes <18yrs | Clinical diagnosis genetically confirmed in ~50% | 31 (% cannot be calculated) | 0.32 | Amed 2010# | |
| Observational investigation of database | Austria / Germany | 262 Pediatric clinics | 40,567 Population | Age <20yrs , Diagnosed <18 yrs | Clinician diagnosis MODY usually confirmed by genetic test (polymorphisms not excluded#) | 339 all cases (0.8%) 263 (0.65%) genetic positive# | Cannot be calculated | Schober 2009 | |
N/K: Not known
only patients with a clinical diagnosis of Type 1 diabetes were included so the prevalence is likely to be underestimated
subsequent study (Awa 2011 ) indicated 38% of reported HNF1A cases were polymorphisms not mutations.
We report the first prevalence study of monogenic diabetes in the UK pediatric population using a systematic screening algorithm and genetic testing for all subtypes of monogenic diabetes.
Aims
To identify the prevalence of monogenic diabetes in the UK pediatric diabetes population by systematic screening.
Methods
Study eligibility
All patients with diabetes who were less than 20 years of age attending one of six pediatric / transition clinics across South West England and Tayside Scotland were eligible to take part. Ethical approval was granted by NRES Committee South West - Central Bristol. Participants under 16 were asked to provide assent and their parents provided consent.
The total number of potential recruits (n=1016) were ascertained by the local pediatric clinical teams from their clinic records i.e. all their patients with diabetes <20 years of age were identified (779 in South West England and 237 in Tayside). Informed consent was obtained by a member of the research team prior to data collection, participants aged 16 or over were asked to provide consent themselves and if they lacked capacity their parents were asked to provide consent. Time from diagnosis was not an exclusion criterion. Data collection included: gender, ethnic group, current age / age at diagnosis, initial / current treatment, time to insulin, family history of diabetes, most recent / highest HbA1c, height / weight at diagnosis and time of recruitment and presence of learning difficulties or deafness. BMI was reported as age adjusted centiles to enable comparison across age groups (28).
Screening method
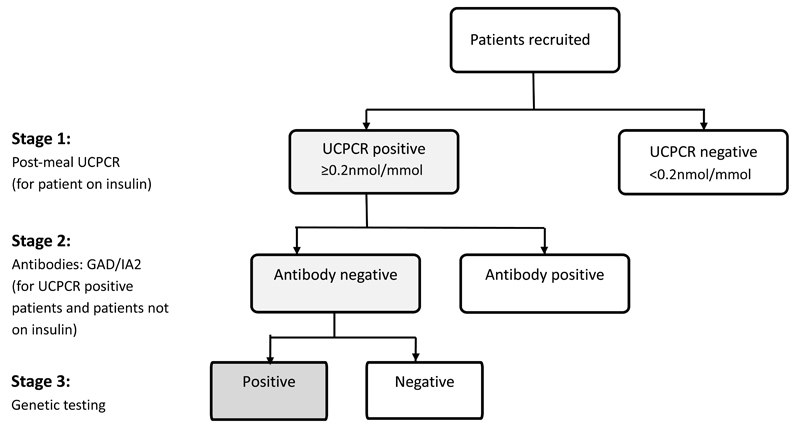
The study comprised of three potential stages which systematically identified those patients eligible for genetic testing (Figure 1).
Figure 1.
Pathway of testing
Stage 1 consisted of a urine sample for measurement of urinary c-peptide creatinine ratio (UCPCR) (14–16). Participants on insulin treatment were asked to mail a urine sample collected two hours after the largest meal of the day that contained carbohydrate to a single laboratory at the Royal Devon and Exeter NHS Foundation Trust. Participants with endogenous insulin production ascertained by UCPCR ≥0.2nmol/mmol and those not on insulin treatment, progressed to stage 2 of the study. Patients with a UCPCR <0.2nmol/mmol, indicating insulin deficiency were considered to have a diagnosis of Type 1 diabetes (14,16).
Stage 2 comprised of a blood sample which tested for the presence of islet auto antibodies (GAD and IA-2) to identify those with autoimmune diabetes. This was performed on all those with significant endogenous insulin (either a UCPCR ≥0.2nmol/mmol on insulin treatment or not on insulin treatment). If islet autoantibody results were available from previous testing, these were used, otherwise a blood sample was taken for antibody testing. Patients with GAD or IA-2 levels >99th centile were deemed islet autoantibody positive (18) and were considered to have a diagnosis of Type 1 diabetes.
Stage 3 consisted of genetic testing in participants who were UCPCR positive and islet autoantibody negative. DNA was extracted, using standard methods, from a blood sample usually taken at the same time as the sample for islet autoantibody testing. Sanger sequencing analysis of the HNF1A and HNF4A genes and dosage analysis by Multiplex Ligation-dependent Probe Amplification (MLPA) to detect partial and whole gene deletions of HNF1A, HNF4A, GCK and HNF1B was undertaken for all patients, with additional Sanger sequencing analysis of the GCK gene undertaken for patients with maximum HbA1c of ≤7.6% (≤60mmol/mol). This testing strategy was performed initially as these are the commonest genes implicated in MODY accounting for >95% of all UK MODY cases (Shields 2010), and are amenable to treatment change. Patients with no pathogenic mutation identified by Sanger sequencing and MLPA then underwent targeted next generation sequencing to look for mutations in 29 genes known to cause monogenic diabetes and the mitochondrial mutation m.3243A>G causing maternally inherited diabetes and deafness using the assay published by Ellard et al (29).
Statistical analysis
Data was double entered onto a database and subsequently cleaned. Data are presented as proportions, and median (IQR) where appropriate, due to non-normality of data. Prevalence was calculated as the proportion of positive cases out of the total number studied. Data was analysed using Stata 13.1.
Results
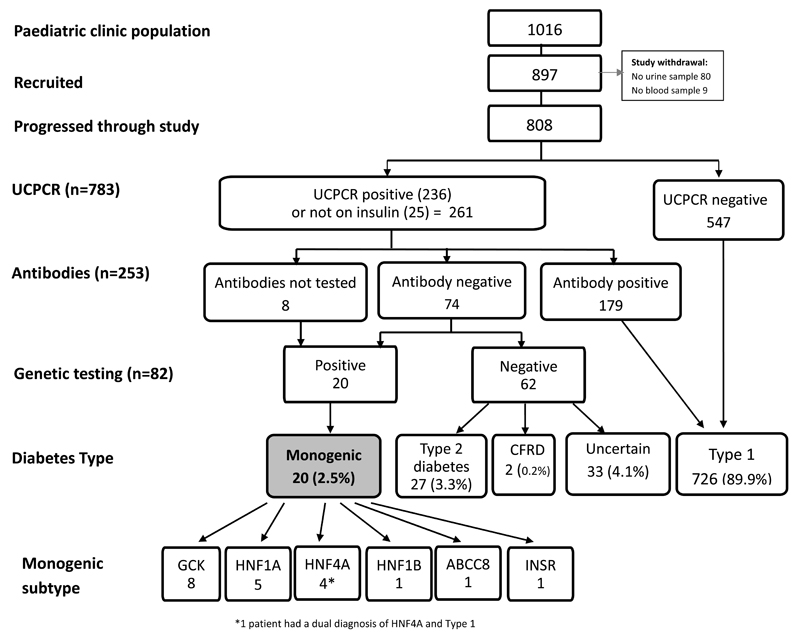
79.5% (n=808/1016) of the eligible population completed the study (Figure 2). 15 of these had previously had genetic testing and were already known to have monogenic diabetes (Table 2).
Figure 2.
Patient progression through pathway
Table 2.
Characteristics of the 20 patients identified with monogenic diabetes
| Study ID | Gene | Mutation | Protein effect | Gender | Age at diagnosis (yrs) | Diabetes duration (yrs)* | Initial treatment | Current treatment | BMI centile | Affected parent | UCPCR nmol/mmol | GAD | IA-2 | Notes |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 211 | GCK | c.97_117dup | p.(Val33_Lys39dup) | M | 3 | 13 | Insulin | None | 99th | Mother | 3.57 | N/A | N/A | Known MODY |
| 537 | GCK | c.683C>T | p.(Thr228Met) | M | 11 | 2 | Diet | None | N/A | Mother | 1.94 | N/A | N/A | Known MODY Sibling of 538 |
| 538 | GCK | c.683C>T | p.(Thr228Met) | M | 9 | 1 | Diet | None | N/A | Mother | 1.73 | N/A | N/A | Known MODY Sibling of 537 |
| 543 | GCK | c.184G>A | p.(Val62Met) | M | 4 | 0.2 | Diet | None | N/A | Mother | N/A | N/A | N/A | Known MODY Sibling of 544 |
| 544 | GCK | c.184G>A | p.(Val62Met) | M | 3 | 2 | Diet | None | N/A | Mother | N/A | N/A | N/A | Known MODY Sibling of 543 |
| 1396 | GCK | c.1209del | p.(Ile404fs) | M | 14 | 0.3 | Diet | None | 71st | Mother | N/A | N/A | N/A | Known MODY |
| 8002095 | GCK | c.1019G>T | p.(Ser340Ile) | M | 9 | 5 | Diet | None | 88th | Father | 0.79 | Neg | N/A | Known MODY |
| 8002372 | GCK | c.1340G>A | p.(Arg447Gln) | M | 18 | 0.6 | Diet | None | 90th | Neither | Not tested | Neg | Not tested | Newly identified MODY |
| 599 | HNF1A | c.608G>A | p.(Arg203His) | F | 14 | 0.5 | OHA | OHA | 99th | Both parents | 3.08 | Neg | Neg | Known MODY |
| 1012 | HNF1A | c.872del | p.(Pro291fs) | F | 10 | 0.7 | Diet | Diet | 99th | Mother | 5.6 | Neg | Neg | Known MODY Sibling of 395 |
| 395 | HNF1A | c.872del | p.(Pro291fs) | F | 14 | 0.1 | OHA | OHA | 95th | Mother | 5.8 | Neg | Neg | Known MODY Sibling of 1012 |
| 455 | HNF1A | c.872dup | p.(Gly292fs) | F | 12 | 3 | OHA | OHA | 57th | Father | 0.86 | Neg | Neg | Known MODY |
| 567 | HNF1A | c.872dup | p.(Gly292fs) | M | 8 | 2 | Diet | OHA | 94th | Mother | 1.73 | Neg | Neg | Known MODY |
| 686 | HNF4A | c.749T>C | p.(Leu250Pro) | M | 16 | 0.7 | Diet | Diet | 99th | Father | 4.74 | N/A | N/A | Known MODY |
| 1348 | HNF4A | c.340C>T | p.(Arg114Trp) | F | 15 | 0.2 | Insulin | OHA | 86th | Father | 3.00 | Neg | Neg | Newly identified MODY |
| 1203 | HNF4A | c.340C>T | p.(Arg114Trp) | M | 7 | 2 | Insulin | Insulin | 39th | Neither | 0.21 | Neg▪ | Neg | Dual diagnosis: Newly identified HNF4A / known Type 1 |
| 377 | HNF4A | c.-12G>A | p.(?) | F | 11 | 2 | Insulin | Insulin | 99th | Mother | 0.28 | Neg | Neg | Newly identified MODY |
| 854 | HNF1B | c.1-?_*151+?del | p.(0?) (whole gene deletion) | M | 11 | 2 | Insulin | Insulin | 9th | Father | 0.71 | Neg | Neg | Newly identified MODY |
| 555 | ABCC8 | c.4139G>A | p.(Arg1380His) | F | 11 | 8 | OHA | OHA | 4th | Father | 3.00 | Neg | Neg | Known MODY |
| 758 | INSR | c.3706C>G | p.(Pro1236Ala) | F | 12 | 3 | OHA | Diet | 55th | Mother | 9.07 | N/A | N/A | Known MODY |
Diabetes duration at time of study
GAD negative as defined in this study as <99th centile, but GAD 25.9 (97.5th centile)
N/A : Not applicable, genetic diagnosis made prior to study
Mutations described using the Human Genome Variation Society (HGVS) nomenclature guidelines according to the following reference sequences: GCK NM_000162.3; HNF1A NM_000545.6; HNF4A NM_175914.4; ABCC8 NM_001287174.1; INSR NM_000208.2
Patient characteristics
54% participants were male (441 male: 376 female). Median age at recruitment was 13 years, [10,16 IQR], median age at diagnosis was 8 years [4,11 IQR] and all individuals were diagnosed with diabetes >6 months of age. Median duration of diabetes was 4.3 years [1.6,7.9 IQR). The majority, 788 (96%) of the cohort were white Caucasian, reflecting the population demographics in these areas. 792 (97%) were insulin treated at time of recruitment, including 4 patients taking insulin in addition to metformin. 25 (3%) patients were non insulin treated with 11 taking oral agents only and 14 on diet alone. Median HbA1c was 8.6% (70 mmol/mol) [7.7,9.7 IQR] (61,83 IQR) and median BMI centile 79 [56,94 IQR ].
Stage 1 - UCPCR
547/817 (67%) were UCPCR negative (<0.2nmol/mol) indicating insulin deficiency and were therefore considered to have Type 1 diabetes and these individuals did not have further testing. 261 (32%) had significant endogenous insulin production (≥0.2nmol/mol); this included 236 patients who were insulin treated and 25 non-insulin treated patients.
Stage 2 - Antibodies
253 patients with significant endogenous insulin underwent islet autoantibody testing, this included 236 patients who were insulin treated and confirmed UCPCR positive through stage 1 of the study and 17 of the non-insulin treated patients. 8/15 patients previously diagnosed with monogenic diabetes did not have antibody testing (but were all non insulin treated)and. 9 patients did not return their blood sample for antibody testing.
179/253 participants were islet autoantibody positive confirming a diagnosis of Type 1 diabetes. 45 of these were positive to both GAD and IA-2, 28 positive to GAD only, 21 positive to GAD but IA-2 not tested and 85 positive to IA-2 only, indicating the importance of testing both autoantibodies. The 74 participants who were antibody negative continued to stage 3 for genetic testing.
Stage 3 - Genetic testing
The prevalence of monogenic diabetes in this UK pediatric diabetes population <20 years was 2.5% (95% CI of 1.5-3.9%). 82/808 (10.1%) patients had genetic testing and 20 (24%) of these (1 in 4) had monogenic diabetes (Table 2). 15/20 patients were previously known to have monogenic diabetes (7 GCK MODY, 5 HNF1A MODY, 1 HNF4A MODY, 1 ABCC8 MODY and 1 patient with type A insulin resistance due to a heterozygous INSR mutation) and 5 new cases of monogenic diabetes (3 HNF4A MODY, 1 HNF1B MODY, 1 GCK MODY) were identified during the study. One of these patients had a dual diagnosis of HNF4A MODY (heterozygous for the p.Arg114Trp mutation) and Type 1 diabetes (GAD negative as defined in this study as <99th centile and therefore proceeded to genetic testing, but GAD titre 25.9 >97.5th centile, UCPCR 0.21nmol/mol two years post diagnosis, on continuous insulin treatment from diagnosis). Cases of monogenic diabetes were found in all 6 clinics with a prevalence varying between 1.2-3.7%.
To assess if we had missed cases of monogenic diabetes in those with islet autoantibodies, 65/179 patients with positive autoantibodies underwent Sanger sequencing analysis of the commonest MODY genes (GCK, HNF1A and HNF4A): no mutations were found.
Characteristics of patients negative on genetic testing
Diagnosis was not established using this testing pathway in 62 participants who were UCPCR positive, islet autoantibody negative and negative for mutations in 29 genes known to cause monogenic diabetes. Secondary causes of diabetes were known in 2 individuals with a previously recorded diagnosis of cystic fibrosis related diabetes. 27/62 of these patients (3.3% of the cohort) met diagnostic criteria for Type 2 diabetes: no monogenic or secondary cause, BMI ≥85th centile and antibody negative (http://public.ukcrn.org.uk/search/StudyDetail.aspx?StudyID=7574) but were not assessed for insulin resistance or other metabolic features.
Uncertainty over the diagnosis remained in 33 individuals (4% of the whole cohort). The most likely diagnosis in these individuals was islet autoantibody negative Type 1 diabetes as they were close to diagnosis (median duration 0.8 years [0.4,2.8 IQR]) and were not overweight (median BMI centile 51st [43,67 IQR]). 26/33 of these had diabetes duration of <3 years, so could be considered within the honeymoon phase and repeating UCPCR in these individuals over time could prove useful. However 5/33 had diabetes duration of median 6.1 years (range 5-10), median BMI 53rd centile (range 46-81) with a UCPCR median 0.36nmol/mmol (range 0.21-1.27) and therefore the diabetes in these individuals should be considered atypical and not fitting a clear diagnostic category.
Only 19.4% (n=198) of the eligible patients within these pediatric diabetes populations did not take part in this study. This included 13 known cases of monogenic diabetes (10 of whom had GCK MODY and were therefore not under the care of a diabetes team, 3 patients with HNF1A and 1 patient with Wolfram Syndrome). Therefore this cohort was not biased to include all those with known monogenic diabetes. The prevalence of monogenic diabetes in those recruited was 2.5% compared to 6.6% (p=0.0038) in those not taking part.
Discussion
We found a prevalence of monogenic diabetes in patients diagnosed under 20 years of 2.5% (95% CI 1.6%, 3.9%) by systematic testing using islet autoantibodies, C-peptide and targeted next generation sequencing of all monogenic diabetes genes. Using our approach of screening children / adolescents with diabetes using C peptide followed by GAD and IA-2 autoantibodies would identify a sub-population of around 10% where genetic testing will have a pick up rate of ~1 in 4. Using the online probability calculator (http://www.diabetesgenes.org/content/mody-probability-calculator) could further aid identification of those most likely to have MODY, as in our study 18/20 with monogenic diabetes were shown to have a 1 in 1.3 chance or >75.5% post test probability of having MODY.
The 2.5% (95% CI 1.6%, 3.9%) prevalence of monogenic diabetes we identified is similar to the three other large systematic population studies: two from predominantly European white populations (Poland (3.1-4.2%), Norway (1.1%)) and one from a multi-ethnic population from the USA (1.4%) (7,2,1, 19) (Table 1). The Polish study used targeted case finding predominantly using clinical criteria supported by the lack of autoantibodies and measurable C peptide. The Norwegian population based study predominantly used antibody negativity combined with a parental history of diabetes or lack of insulin therapy and HbA1c<7.5% (58mmol/mol) (2). The lower prevalence (1.1%) probably reflected that they studied children aged 0-14 years rather than 0-20 years (mean age of diagnosis 10.6 years in our study) and only 10 patients were tested for glucokinase. The US study, like our study, used systematic biomarker screening with genetic testing performed in all patients who had measurable C peptide and did not have GAD and IA-2 autoantibodies. The lower prevalence in their cohort probably results from non MODY patients having more ‘Type 2 features’ suggesting a greater proportion of patients with young onset Type 2 diabetes, as the combined prevalence of MODY in minorities was very similar to the prevalence of MODY in non-Hispanic whites (1). There are many other less comprehensive studies of the prevalence of monogenic diabetes (Table 1): these are limited by studying a single clinic, using a non-systematic assessment and/or not making a robust molecular genetic diagnosis (confirmed mutations not polymorphisms) (25,27,30).
Our study indicated a higher proportion of known cases of MODY versus new cases identified through systematic screening. The 28 cases of previously confirmed MODY (15 who took part and 13 who did not take part in the study) reflects the high levels of awareness of monogenic diabetes in these geographical regions. The 13 cases previously identified who did not take part included 9 with GCK MODY (who had been discharged from clinic follow up), 3 with HNF1A MODY and 1 with Wolfram Syndrome. This study shows that clinical recognition of key phenotypes in the cases and their family members can identify the majority of paediatric patients: (15/20) in this study. However the 5 new cases identified through this pathway of screening indicates the need for a systematic approach. If this approach was used in other areas where recognition of monogenic diabetes is not so apparent then a greater proportion of new cases would be identified. We have based our prevalence figures only on the population recruited in this study, however if the 13 patients who did not take part were taken into account this could give a prevalence as high as 3.3% (33/1016). There are estimated to be around 35,000 children and young people with diabetes, under the age of 19, in the UK (31,32). If the prevalence of 2.5% found in those who took part in this 6 clinic survey reflects the whole of the UK then this suggests at least 875 (95% CI 560-1365) expected cases of MODY in this age group, of whom 468 have been diagnosed to date with approximately 50% still likely to be misdiagnosed with Type 1 diabetes.
This approach of systematic testing combined with clinical criteria can result in a diagnosis in over 99% of cases and this is an advantage of this approach above the recognition of monogenic diabetes. We were able to use C peptide, autoantibody and genetic testing to give a clear diagnosis in 92.3% of cases. Clinical criteria suggests that 3.3% had Type 2, a figure very similar to the amount of monogenic diabetes, as seen in other European populations (7, 24, 33). 0.2% had secondary diabetes due to cystic fibrosis which probably reflects an underestimate as many of these patients will not attend a paediatric diabetes clinic. A further 3.2% were within three years of diagnosis and were probably antibody negative Type 1 diabetes in the honeymoon period. There remained 5 patients (0.6%) who are atypical and hard to classify – they may represent atypical Type 1 diabetes (antibody negative and significant C peptide more than 3 years after diagnosis) or a presently unrecognised subtype of monogenic diabetes.
There were limitations to this study. The geographical areas where the study was undertaken already had a high awareness of MODY so the number of new cases was low (25%) relative to those already known (75%) while elsewhere in the UK we estimate this figure is approximately 50% detected and 50% undetected. We only systematically genetically tested patients who had significant endogenous insulin (C peptide) and did not have autoantibodies although previous research (14,17,18) and our failure to find any mutations in 65 patients who did have significant C peptide but were antibody positive supports that this approach would miss very few cases. UCPCR was conducted irrespective of duration of diabetes and it is acknowledged that some of the patients tested close to diagnosis could be producing endogenous insulin during the honeymoon period and if retested over time those with Type 1 diabetes would be expected to have declining c-peptide levels. UCPCR is best for excluding patients 3 years after diabetes is diagnosed while autoantibodies are best close to diagnosis. In this study we wanted to test everyone irrelevant of disease duration so a method that used both biomarkers worked well. If a study was performed of purely incident cases, which would have an advantage of making the correct diagnosis early, then measuring C peptide would have little value and further testing could be performed on those that were negative to multiple autoantibodies. Although patients were asked to send a ‘post meal’ urine sample the prandial state of the patient was assumed (and not observed) and therefore we cannot be certain that all UCPCR tests were stimulated. Our population were predominantly (96%) white Caucasians and systematic studies in other, especially high prevalence, populations are also needed.
There are many strengths of this study. The result is likely to be representative of the clinics studied as 79.5% of the eligible population took part; a result that shows the high acceptability of this approach in paediatric clinics. The systematic biomarker based approach that is independent of clinical features allows atypical cases to be detected (e.g. those with no family history of diabetes). This is the first study that has used next generation sequencing to assess all known causes of monogenic diabetes although the majority (85%) had the most common MODY genes GCK, HNF1A and HNF4A so studies that have not used this approach will have only missed a few patients.
Conclusion
This systematic, high uptake study gives a prevalence of 2.5% (95% CI 1.6%, 3.9%) of monogenic diabetes in the UK pediatric population. Patients with monogenic diabetes were identified in every pediatric clinic. The successful identification of patients with monogenic diabetes is crucial as they require different treatment than Type 1 or Type 2 diabetes. The vast majority (>99%) of pediatric patients can be successfully classified by UCPCR, antibody testing, genetic testing and clinical criteria. UCPCR is a non-invasive and inexpensive test which could be more widely used in the paediatric age group where it has a high acceptability. This screening algorithm is a practical approach to determining the prevalent cases in a clinic to ensure correct diagnosis of subtypes of diabetes. Confirming a prevalence of MODY of 2.5% in the pediatric population indicates that all those involved in pediatric diabetes care should be aware of the possibility of an alternative diagnosis and know how to refer patients for genetic testing.
Acknowledgements
We are grateful to the patients who took part in this study and the nurses who recruited them for their support with this project.
This work presents independent research commissioned by the Health Innovation Challenge Fund [grant number HICF-1009-041], a parallel funding partnership between the Wellcome Trust and the Department of Health and was supported by the National Institute for Health Research (NIHR) Exeter Clinical Research Facility and the SW Peninsula Diabetes Research Network. MS is supported by the NIHR Exeter Clinical Research Facility. ERP is a Wellcome Trust New Investigator. ATH and SE are both Wellcome Trust Senior Investigators. ATH is an NIHR Senior Investigator. TM is funded by an NIHR CSO Fellowship. The views expressed in this publication are those of the author(s) and not necessarily those of the Wellcome Trust, the NHS, the NIHR or the Department of Health.
Footnotes
Author contributions
MS wrote the manuscript, collected and analysed the data. BS analysed the data and reviewed / edited the manuscript. SH collected the data and reviewed the manuscript. MH co-ordinated the project, assisted with the data and reviewed the manuscript, TJM co-ordinated the UCPCR and antibody testing and reviewed the manuscript. KC assisted with genetic data and reviewed / edited the manuscript, RO contributed to the discussion and reviewed the manuscript , BK developed the protocol, submitted the ethics application and reviewed the manuscript, CH reviewed the manuscript, JC, KM, CM, RS, BF, SR, and SG facilitated patient recruitment within their pediatric clinics and reviewed the manuscript, SE co-ordinated the genetic testing and reviewed the manuscript, ERP co-ordinated the Tayside arm of the project and reviewed / edited the manuscript, ATH designed the study, contributed to the discussion, reviewed and edited the manuscript, the UNITED team collected the data. MS is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
There are no conflicts of interest to declare.
Diabetes care
This is an author-created, un-copy-edited electronic version of an article accepted for publication in Diabetes Care. The American Diabetes Care Association (ADA), publisher of Diabetes Care, is not responsible for any errors or omissions in this version of the manuscript or any version derived from it by third parties. The definitive publisher-authenticated version will be available in a future issue of Diabetes Care in print and online at http://care.diabetesjournals.org,
References
- 1.Pihoker C, Gilliam LK, Ellard S, et al. SEARCH for Diabetes in Youth Study Group. Prevalence, characteristics and clinical diagnosis of maturity onset diabetes of the young due to mutations in HNF1A, HNF4A and glucokinase: results from the SEARCH for diabetes in youth. J Clin Endocrinol Metab. 2013;98(10):4055–4062. doi: 10.1210/jc.2013-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Irgens HU, Molnes J, Johansson BB, et al. Prevalence of monogenic diabetes in the population based Norwegian childhood diabetes registry. Diabetologia. 2013;56:1512–1519. doi: 10.1007/s00125-013-2916-y. [DOI] [PubMed] [Google Scholar]
- 3.Carmody D, Lindauer KL, Naylor RN. Adolescent non-adherence reveals a genetic cause for diabetes. Diabet Med. 2015 Jun;32(6):e20–3. doi: 10.1111/dme.12669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gandica RG, Chung WK, Deng L, Goland R, Gallagher MP. Identifying monogenic diabetes in a pediatric cohort with presumed type 1 diabetes. Pediatr Diabetes. 2015 May;16(3):227–33. doi: 10.1111/pedi.12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lambert AP, Ellard S, Allen LI, et al. Identifying hepatic nuclear factor 1 alpha mutations in children and young adults with a clinical diagnosis of type 1 diabetes. Diabetes Care. 2003;26:333–337. doi: 10.2337/diacare.26.2.333. [DOI] [PubMed] [Google Scholar]
- 6.Thirumalai A, Holing E, Brown Z, Gilliam LK. A case of hepatocyte nuclear factor-1β (TCF2) maturity onset diabetes of the young misdiagnosed as type 1 diabetes and treated unnecessarily with insulin. J Diabetes. 2013 Dec;5(4):462–4. doi: 10.1111/1753-0407.12043. [DOI] [PubMed] [Google Scholar]
- 7.Fendler W, Borowiec M, Baranowska-Jazwiecka A, et al. Prevalence of monogenic diabetes amongst Polish children after a nationwide genetic screening campaign. Diabetologia. 2012;55:2631–2635. doi: 10.1007/s00125-012-2621-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rubio-Cabezas O, Edghill EL, Argente J, Hattersley AT. Testing for monogenic diabetes among children and adolescents with antibody negative clinically defined Type 1 diabetes. Diabetic Medicine. 2009;26:1070–1074. doi: 10.1111/j.1464-5491.2009.02812.x. [DOI] [PubMed] [Google Scholar]
- 9.Murphy R, Ellard S, Hattersley AT. Clinical implications of a molecular genetic classification of monogenic beta-cell diabetes. Nat Clin Pract Endocrinol Metab. 2008 Apr;4(4):200–13. doi: 10.1038/ncpendmet0778. [DOI] [PubMed] [Google Scholar]
- 10.Rubio-Cabezas O, Hattersley AT, Njølstad PR, et al. The diagnosis and management of monogenic diabetes in children and adolescents. Pediatric Diabetes. 2014;15(Suppl. 20):47–64. doi: 10.1111/pedi.12192. [DOI] [PubMed] [Google Scholar]
- 11.Craig ME, Jefferies C, Dabelea D, et al. ISPAD clinical practice consensus guidelines 2014 compendium. Definition, epidemiology and classification of diabetes in children and adolescents. Pediatric Diabetes. 2014;15(Suppl 20):4–17. doi: 10.1111/pedi.12186. [DOI] [PubMed] [Google Scholar]
- 12.Colclough K, Saint-Martin C, Timsit J, Ellard S, Bellanné-Chantelot C. Clinical utility gene card for: Maturity-onset diabetes of the young. European Journal of Human Genetics. 2014;22 doi: 10.1038/ejhg.2014.14. 10.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shields BM, Hicks S, Shepherd MH, et al. Maturity onset diabetes of the young (MODY): how many cases are we missing ? Diabetologia. 2010;53:2504–2508. doi: 10.1007/s00125-010-1799-4. [DOI] [PubMed] [Google Scholar]
- 14.Besser RE, Shepherd MH, McDonald TJ, et al. Urinary C-peptide creatinine ratio is a practical outpatient tool for identifying hepatocyte nuclear factor 1-{alpha}/hepatocyte nuclear factor 4-{alpha} maturity-onset diabetes of the young from long-duration type 1 diabetes. Diabetes Care. 2011 Feb;34(2):286–91. doi: 10.2337/dc10-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McDonald TJ, Knight BA, Shields BM, et al. Stability and reproducibility of a single-sample urinary C-peptide/creatinine ratio and its correlation with 24-h urinary C-peptide. Clin Chem. 2009 Nov;55(11):2035–9. doi: 10.1373/clinchem.2009.129312. [DOI] [PubMed] [Google Scholar]
- 16.Besser RE, Ludvigsson J, Jones AG, et al. Urine C-peptide creatinine ratio is a non-invasive alternative to the mixed-meal tolerance test in children and adults with type 1 diabetes. Diabetes Care. 2011 Mar;34(3):607–9. doi: 10.2337/dc10-2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Besser RE, Shields BM, Hammersley SE, et al. Home urine C-peptide creatinine ratio (UCPCR) testing can identify type 2 and MODY in pediatric diabetes. Pediatr Diabetes. 2013 May;14(3):181–8. doi: 10.1111/pedi.12008. [DOI] [PubMed] [Google Scholar]
- 18.McDonald TJ, Colclough K, Brown R, et al. Islet autoantibodies can discriminate maturity-onset diabetes of the young (MODY) from Type 1 diabetes. Diabet Med. 2011 Sep;28(9):1028–33. doi: 10.1111/j.1464-5491.2011.03287.x. [DOI] [PubMed] [Google Scholar]
- 19.Shankar RK, Pihoker C, Dolan LM, et al. Permanent neonatal diabetes mellitus: prevalence and genetic diagnosis in the SEARCH for diabetes in youth study. Pediatr Diabetes. 2013;14(3):174–180. doi: 10.1111/pedi.12003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hameed S, Ellard S, Woodhead HJ, et al. Persistently autoantibody negative (PAN) type 1 diabetes mellitus in children. Pediatric Diabetes. 2011;12:142–149. doi: 10.1111/j.1399-5448.2010.00681.x. [DOI] [PubMed] [Google Scholar]
- 21.Wheeler BJ, Patterson N, Love DR, et al. Frequency and genetic spectrum of maturity onset diabetes of the young (MODY) in southern New Zealand. Journal of Diabetes and Metabolic Disorders. 2013;12:46. doi: 10.1186/2251-6581-12-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Redondo MJ, Rodriguez LM, Escalante M, et al. Types of pediatric diabetes mellitus defined by anti-islet autoimmunity and random C-peptide at diagnosis. Pediatric Diabetes. 2013;14:333–340. doi: 10.1111/pedi.12022. [DOI] [PubMed] [Google Scholar]
- 23.Ehtisham S, Hattersley A, Dunger D, Barrett T. First UK survey of pediatric type 2 diabetes and MODY. Arch Dis Child. 2004;89(6):526–529. doi: 10.1136/adc.2003.027821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neu A, Feldhahn L, Ehehalt S, Hub R, Ranke M. Type 2 diabetes mellitus in children and adolescents is still a rare disease in Germany: a population based assessment of the prevalence of type 2 diabetes and MODY patients aged 0-20 years. Pediatric Diabetes. 10:468–473. doi: 10.1111/j.1399-5448.2009.00528.x. [DOI] [PubMed] [Google Scholar]
- 25.Amed S, Dean HJ, Panagiotopoulos C, et al. Type 2 diabetes, medication induced diabetes and monogenic diabetes in Canadian children. Diabetes Care. 2010;33(4):786–791. doi: 10.2337/dc09-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galler A, Stange T, Müller G, et al. for the childhood diabetes registry in Saxony, Germany. Incidence of childhood diabetes in children aged less than 15 years and its clinical and metabolic characteristics at the time of diagnosis: data from the childhood diabetes registry of Saxony, Germany. Horm Res Paediatri. 2010;74:285–291. doi: 10.1159/000303141. [DOI] [PubMed] [Google Scholar]
- 27.Schober E, Rami B, Grabert M, et al. Phenotypical aspects of maturity onset diabetes of the young (MODY diabetes) in comparison with type 2 diabetes mellitus (T2DM) in children and adolescents: experience from a large multicentre database. Diabet Med. 2009;26:466–473. doi: 10.1111/j.1464-5491.2009.02720.x. [DOI] [PubMed] [Google Scholar]
- 28.Freeman JV, Cole TJ, Chinn S, et al. Cross sectional stature and weight reference curves for the UK, 1990. Arch Dis Child. 1995 Jul;73(1):17–24. doi: 10.1136/adc.73.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ellard S, Lango Allen H, De Franco E, et al. Improved genetic testing for monogenic diabetes using targeted next-generation sequencing. Diabetologia. 2013 Sep;56(9):1958–63. doi: 10.1007/s00125-013-2962-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Awa WL, Thon A, Raile K, et al. Genetic and clinical characteristics of patients with HNF1A gene variations from the German-Austrian DPV database. European Journal of Endocrinology. 2011;164:513–520. doi: 10.1530/EJE-10-0842. [DOI] [PubMed] [Google Scholar]
- 31.HSCIC. National Diabetes Audit 2011\12: Report 1: Care Processes and Treatment Targets. http://www.hscic.gov.uk/searchcatalogue?productid=13129&q=%22National+diabetes+audit%22&sort=Relevance&size=10&page=1#top. (29,576 registered with GP practices within the NDA survey)
- 32.Scottish Diabetes Survey. 2012 http://www.diabetesinscotland.org.uk/Publications.aspx. (3,827)
- 33.Vaziri-Sani Fariba, Delli Ahmed J, Elding-Larsson Helena, et al. on behalf of the Swedish Better Diabetes Diagnosis Study Group A novel triple mix radiobinding assay for the three ZnT8 (ZnT8-RWQ) autoantibody variants in children with newly diagnosed diabetes. J Immunol Methods. 2011 Aug 31;371(1-2):25–37. doi: 10.1016/j.jim.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yorifuji T, Fujimaru R, Hosokawa Y, et al. Comprehensive molecular analysis of Japanese patients with pediatric-onset MODY type diabetes mellitus. Pediatric Diabetes. 2012;13:26–32. doi: 10.1111/j.1399-5448.2011.00827.x. [DOI] [PubMed] [Google Scholar]
- 35.Chambers C, Fouts A, Dong F, et al. Characteristics of maturity onset diabetes of the young in a large diabetes center. Pediatric Diabetes. 2015 Jun 8; doi: 10.1111/pedi.12289. [DOI] [PMC free article] [PubMed] [Google Scholar]