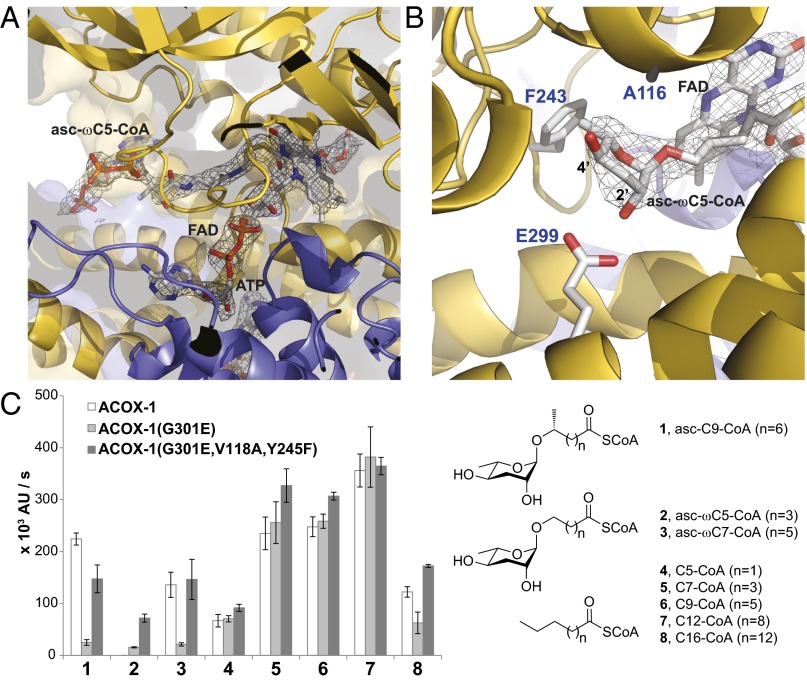
Fig. 2.
Comparison of the active sites of ACOX-1 and ACOX-2. (A) Active site of ACOX-2(E432A) with bound asc-ωC5-CoA (2) substrate and FAD cofactor. The electron density associated with the substrate and cofactor is shown (2Fo-Fc map, σ=1.0). (B) Close up of the asc-ωC5-CoA (2) substrate in the active site of ACOX-2(E432A), shown from the opposite perspective as shown in A. Glu-299 hydrogen bonds to the 2′-hydroxyl of the ascarylose ring of the substrate. Phe-243 and Ala-116 help to accommodate the ascarylose ring. (C) In vitro enzyme activity assay showing the effect of mutating active site residues (Gly-301, Val-118, and Tyr-245) of ACOX-1 to the corresponding residues in ACOX-2. The structures of the (ω-1)-ascaroside-CoA (1), ω-ascaroside-CoA (2–3), and fatty acyl-CoA (4–8) substrates that were tested are shown. Data represent the mean ± SD of three independent experiments.