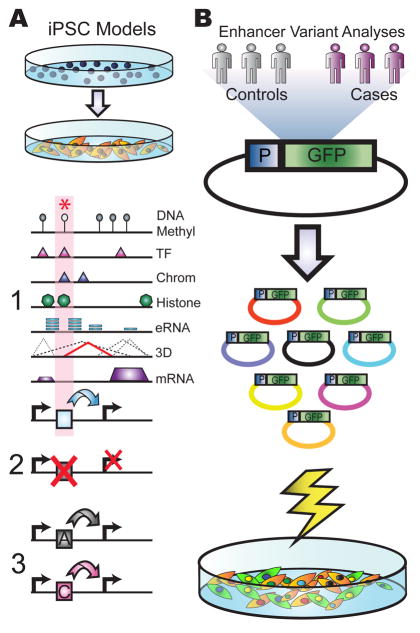
Figure 2. Next-generation sequencing based disease studies.
Experimental approaches that can be applied for assessing the role of enhancers or other non-coding, regulatory variants in diseases are shown. (A) A schematic of patient-derived iPSC models is given. Reprogrammed patient iPSCs into relevant cell types can be used for functional genomic studies and cellular phenotypic analyses. These functional genomic studies can identify key elements using an integrative approach that incorporates complementary information, including DNA methylation (DNA Methyl), DNA-binding protein occupancy (TF), regions of open chromatin (Chrom), histone modifications (Histone), eRNA production (eRNA), long range interactions (3D) and RNA expression (mRNA) to identify regulatory elements (1). Subsequent CRISPR genome editing of key regulatory elements is used to validate target genes (2), whereas regulatory element swapping can be used to predict the functional effect of regulatory sequence variants (3). (B) A high-throughput reporter assay screen is shown. After construction and transfection of a complex pool of reporter plasmids harboring hundreds of enhancer sequences from case and control populations, NGS can identify both rare and common variants within enhancer sequences that lead to functional effects on gene expression. (P = Promoter; GFP = Green Fluorescent Protein).