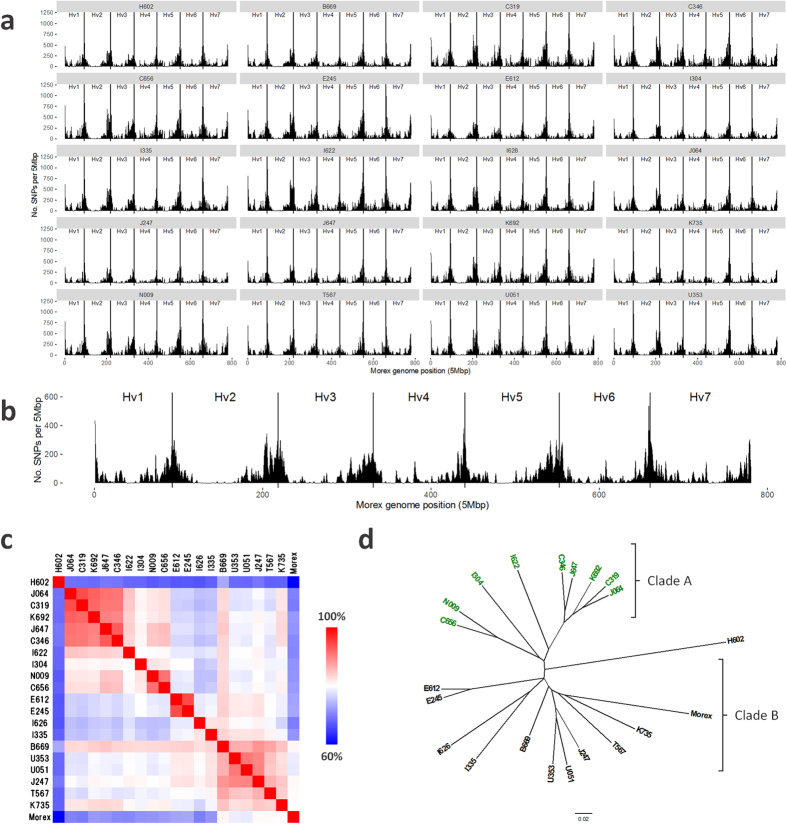
Figure 3. Genome-wide distribution of the SNPs identified using RNA-seq analysis of the 20 diverse barley accessions.
(a) Distribution of the RNA-seq-based SNPs of the 20 barley accessions, using the Morex genome as a reference. The numbers of SNPs were computed for each chromosome using 5 million bases sliding windows. (b) Distribution of the bi-allelic RNA-seq-based SNPs. (c) Genome-wide sequence identity based on the bi-allelic SNP dataset across the 20 barley accessions and Morex. (d) Neighbour-joining tree of the 20 barley accessions and Morex based on the bi-allelic SNP dataset.