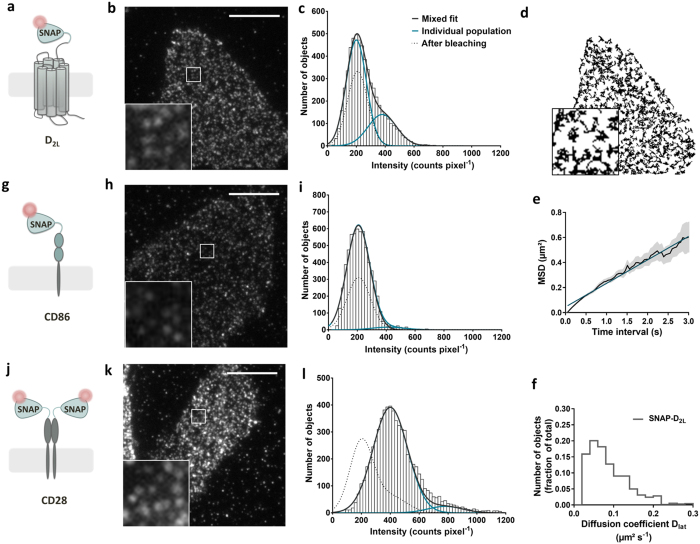
Figure 1. Visualization, tracking and analysis of the dimerization of single SNAP-tagged D2L receptors using SNAP-CD86 and SNAP-CD28 as monomeric and dimeric reference proteins.
(a,g,j) Schematic representation of the SNAP-tagged constructs. (b,h,k) Representative images of single CHO cells, stably transfected with the corresponding labeled protein and visualized by TIRF-M. Scale bar, 10 μm. The first 100 frames of the cell in b are shown in Supplementary Movie S1. Inserts correspond to higher magnification images of the areas in the white boxes. (c,i,l) Representative intensity distributions of fluorescent spots identified over the first 10-frame time window of TIRF illumination of CHO cells, stably transfected with the corresponding construct and labeled with Alexa546-BG. Number of identified particles, n = 5770 (c), 6252 (f) and 6458 (i). Data were fitted with a mixed Gaussian model. A mixed Gaussian fit after partial photobleaching (dotted line) was used to estimate the intensity of a single fluorescent molecules in each image sequence. (d) Individual trajectories of moving SNAP-D2L receptors were identified from the entire recording of the cell shown in (b). The insert shows a higher magnification that illustrates the random nature of the diffusive process. (e) Representative plot of the average mean square displacement (MSD) (mean ± s.d.) versus the time interval (δt) for the trajectories shown in (a). The plot is linear (r2 = 0.99 – linear fit (blue)), over a 3-s timescale, which is consistent with receptor movement following a random walk, and it shows no evidence for anomalous diffusive behavior. (f) Distribution of the diffusion coefficients of the receptor particles tracked in (d).