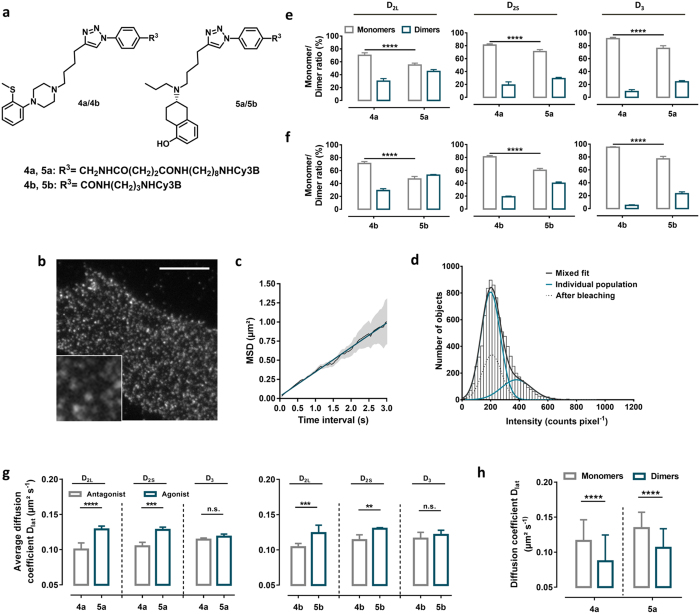
Figure 8. Influence of antagonists (4a,b) and agonists (5a,b) on D2L, D2S and D3 receptor dimerization.
(a) Chemical structures of monovalent fluorescent antagonists (4a,b) and agonists (5a,b). (b) Representative images a single CHO cell stably expressing the D2L receptor, labeled with 4a (38 nM), visualized by TIRF-M. Scale bar, 10 μm. Insert correspond to a higher magnification image of the area in the white box. (c) Plot of average mean square displacement (MSD ± s.d.) versus the time interval (δt) of the receptor ligand complexes that were tracked (r2 = 0.99 – linear fit (blue)). (d) Intensity distribution of fluorescent spots identified over the first 10-frame time window of ligand receptor complex D2L-4a. (e,f) Monomer/dimer ratios of D2L, D2S and D3 receptors labeled with the fluorescent antagonists 4a,b and agonists 5a,b, calculated from fitted fluorescence intensity distributions of fluorescent ligand receptor complexes using a mixed Gaussian model (compare to (c) and Supplementary Table S5). (g) Average diffusion coefficients (Dlat) of the receptor complexes of the same cells analyzed in (e,f). Data in (e–g) represent mean ± s.d. n analysed cells (D2L: n = 18 for 4a, 19 for 5a, 14 for 4b and 17 for 5b, D2S: 13 for 4a, 12 for 5a, 19 for 4b and 12 for 5b, D3: 12 for 4a, 13 for 5a, 15 for 4b and 10 for 5b). (e) Representative effects of the size of D2L receptor-ligand complexes on their rates of lateral diffusion. The diffusion coefficient of D2L-4a monomers (0.116 ± 0.030 μm2 s−1, n = 838 from 3 cells) is greater than that of the dimers (0.087 ± 0.037 μm2 s−1, n = 185 from 3 cells). This is also found for D2L-5a monomers 0.134 ± 0.023 μm2 s−1 (n = 1396 from 3 cells) and dimers 0.106 ± 0.027 μm2 s−1 (n = 558 from 3 cells). Data represent means ± s.d. Statistical analysis was performed by an unpaired t-test (**p-value < 0.01, ***p-value < 0.001, ****p-value < 0.0001, n.s. - not significant).