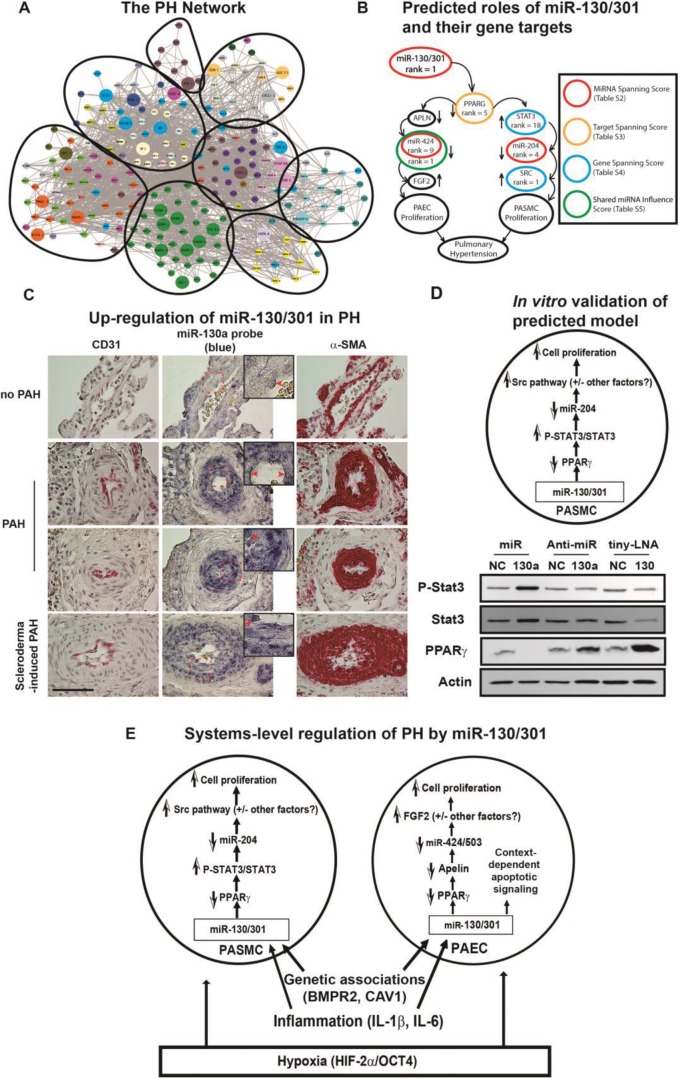
Figure 1.
Prediction of microRNA (miR-)130/301-PPARγ (peroxisome proliferator–activated receptor γ; also PPARG) signaling axis in pulmonary hypertension (PH) via in silico network modeling. A, As we previously reported,1 we constructed a comprehensive network of pulmonary hypertension (PH)–relevant genes and analyzed its pathway enrichment (color coding denotes functional pathway) and architectural structure (circles denote architectural clusters). B, We ranked microRNAs (miRNAs/miRs) on the basis of the size and intercluster spread of their target pools and identified miR-130/301 as a highly influential regulatory factor. We ranked genes in the miR-130/301 target pool in silico by “hubness” and other centrality metrics and found top-ranking genes to form the outline of two known proliferative pathways in the lung. C, D, We verified these findings in vivo and in vitro, demonstrating that miR-130/301 was upregulated in the pulmonary vasculature of PH patients (multiple etiologies; C) and that it modulated vascular proliferation in PAECs (data not shown) and PASMCs by the STAT3-miR-204-SRC axis (D). E, As predicted by our network-based algorithms, a model was confirmed of the global influence of miR-130/301 on vascular proliferation in response to a variety of PH triggers. α-SMA: α smooth muscle actin; APLN: apelin; BMPR2: bone morphogenetic protein receptor 2; CAV1: caveolin 1; FGF2: fibroblast growth factor 2; HIF-2α: hypoxia-inducible factor 2α; IL: interleukin; OCT4: octamer-binding transcription factor 4; PAEC: pulmonary arterial endothelial cell; PAH: pulmonary arterial hypertension; PASMC: pulmonary arterial smooth muscle cell; STAT3: signal transducer and activator of transcription 3. Images adapted from Bertero et al.1 and reprinted with permission.