Table 1. Alternative splicing implied by putative novel introns (PNIs).
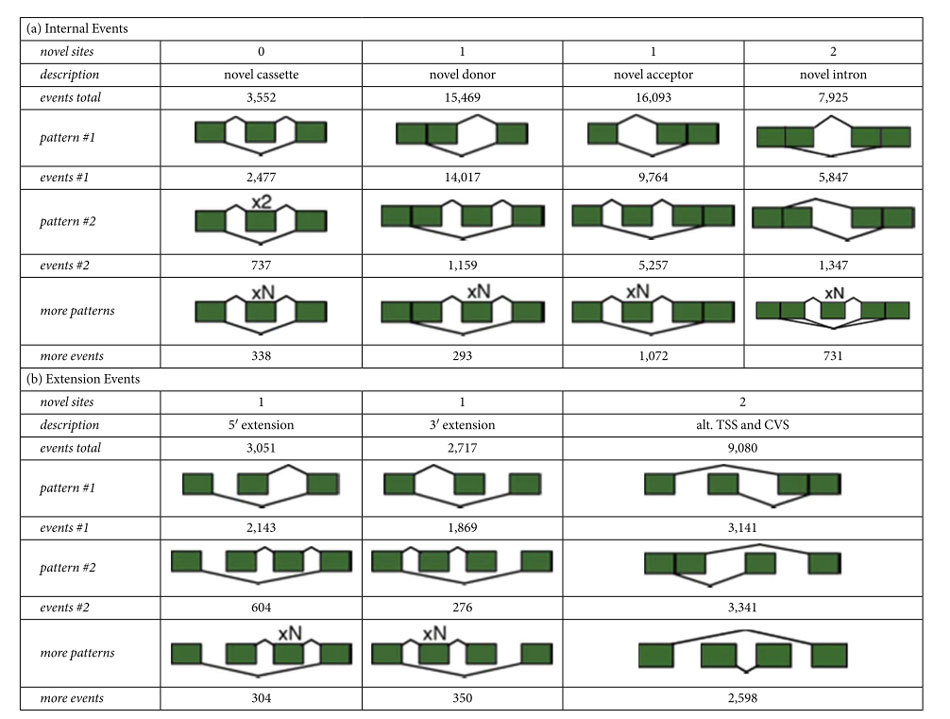
The table summarises novel alternative splicing events implied by superimposing the 21,761 Gencode PNIs supported by >150 individuals to the transcript structures of the Gencode reference annotation. The events have been grouped according to their localisation within the transcript body (i.e., “internal events”, Table 1a) or beyond the transcript extremities (“extension events”, Table 1b). The 1st row presents the number of novel splice sites, i.e. the splice sites of the PNI that are not annotated in the Gencode reference, described in each category (i.e., the column). The 2nd row provides the total count of such events. Rows 3–8 show the two most frequently observed event patterns (“pattern #1” and “pattern #2”) in the category and a summary of remaining patterns (“more patterns”), with the corresponding number of single events observed for each pattern. (a) Most internal PNIs link novel splice sites to an existing one (~73%), less frequently introns employ two novel sites (~18%), and novel combinations of existing sites are rather exceptional (~8%). (b) In contrast, PNIs employing novel splice sites upstream of the annotated transcription start site (TSS) or downstream of the annotated cleavage site (CVS) are more frequently combinations of two novel splice sites (~62% vs. ~59%).
