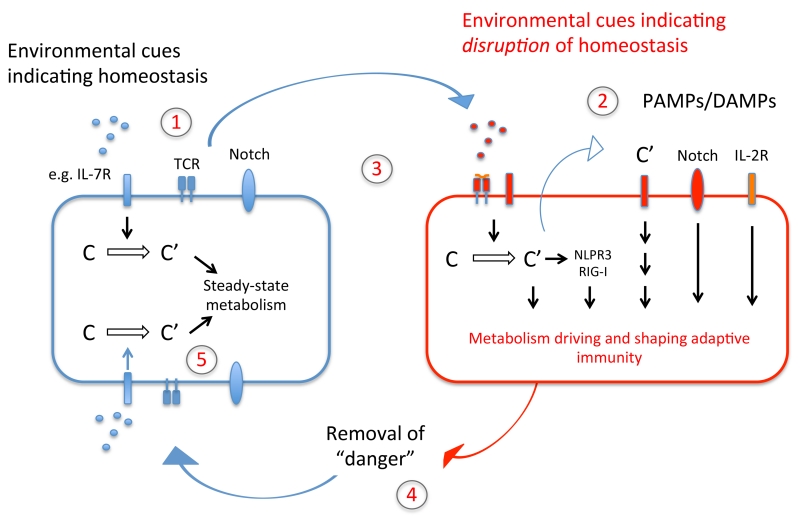
Figure 4. ‘Feedback model’ of the complement–metabolism system in T cells.
During organismal homeostasis, intracellularly generated complement fragments sustain T cell self-regulation through the fine-tuning of basal metabolic activity, and this quiescent state is supported by complement driven assembly of the IL-7R and restraining of Notch activity (1). Together with cognate T cell activation, signals indicative of disrupted homeostasis (2), initiate complement-driven metabolic reprogramming, IL-2R assembly, Notch and NLRP3 inflammasome activation and context-specific activation of the cell (3). This enables an efficient immune response, which in turn closes a feedback-loop to the sensing system by modifying the input on the sensor-module – namely removal of cognate pathogen and thus the inflammatory trigger (4). Metabolic reprogramming back towards steady-state metabolism thus occurs (5). The same basic principle of a complement-dependent ‘sensing–response–feedback system’ would also apply to other immune cells (for example, antigen-presenting cells, where TLR activation may induce complement-mediated metabolic programming) and in cell-autonomous models, e.g. when intracellular sensing of C3 fragments deposited onto cell-invading pathogens triggers a mitochondrial antiviral signaling protein-driven immune response, leading to degradation of the pathogens by the proteasome. Of note, we have predominantly integrated into this schematic the effector systems networking with complement that have a defined metabolic role in immune cell activation.