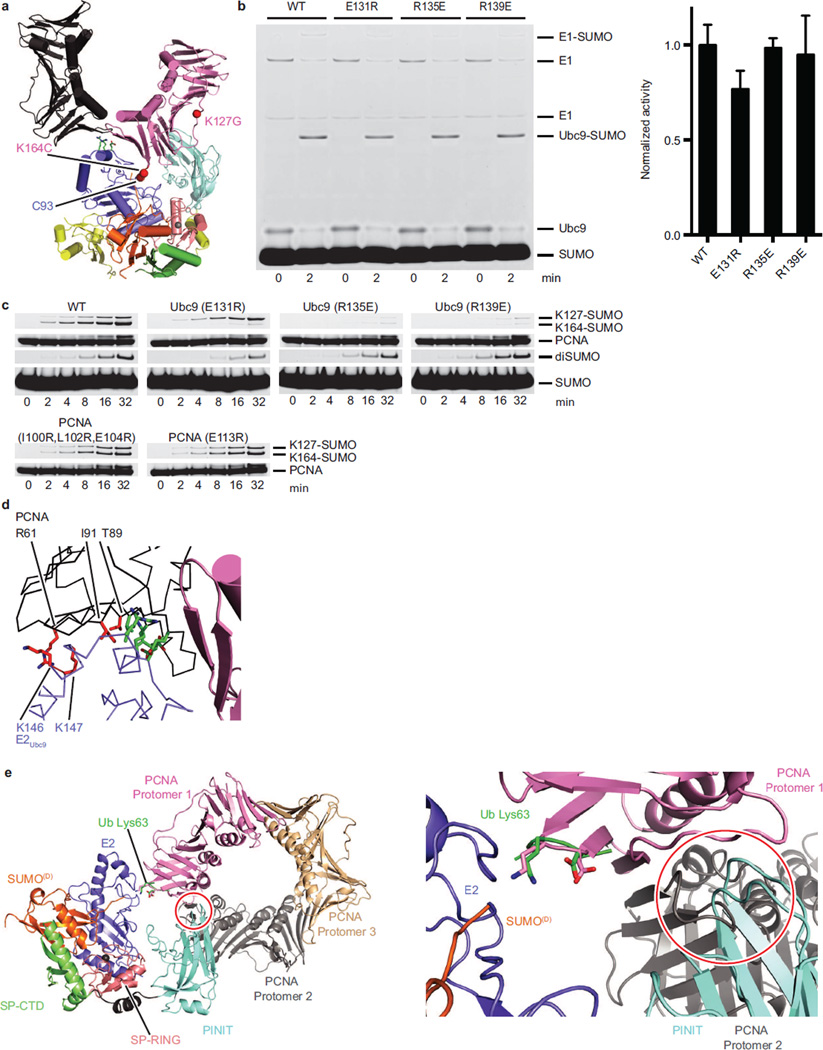
Extended Data Figure 6. Shape Complementarity between the E2Ubc9-SUMO/E3 Complex and PCNA.
a, The current structure (color) with the crystallographic packing of a lattice mate PCNA molecule (black). b, Non-reducing SDS-PAGE analysis of 2 minute endpoint in vitro E2Ubc9-SUMO thioester formation reactions with 0.05 µM E1, 0.4 µM of the indicated E2Ubc9 and 22 µM SUMO (left) and the quantitated E2-SUMO band (right). The quantified band intensity shows mean ± s.d. (n=3 technical replicates). c, SDS-PAGE analysis of multiple turnover assays of SUMO modification of PCNA utilizing in vitro reactions with coupled E1 (200 nM), E2 (100 nM), and E3 (50 nM) activities with 4 µM PCNA or without PCNA (diSUMO formation) shown quantified in Fig. 5c. d, Location of E2Ubc9 and PCNA mutations that had no effect (red sticks) on activities observed for in vitro assays similar to those performed in Fig. 5c in relation to residues that did show effects (green sticks). e, The E2Ubc13C87S-Ub was aligned to E2Ubc9 in the current structure and subsequently the Lys164/Glu165 loop from trimeric PCNA (pink) was aligned onto the Lys63/Glu64 loop from acceptor ubiquitin (2GMI, green). Within this conformation the E3Siz1 PINIT domain (cyan) clashes with another protomer of the PCNA trimer (grey). For gel source data, see Supplementary Figure 1.