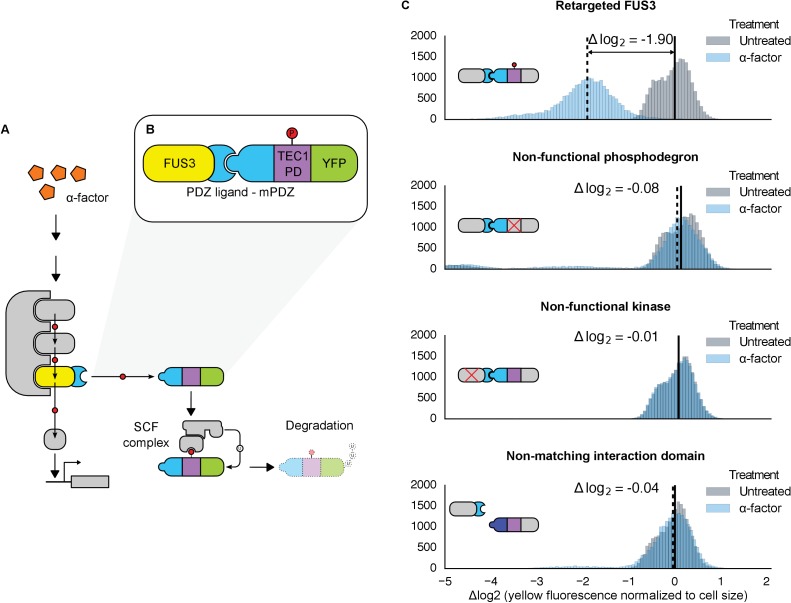
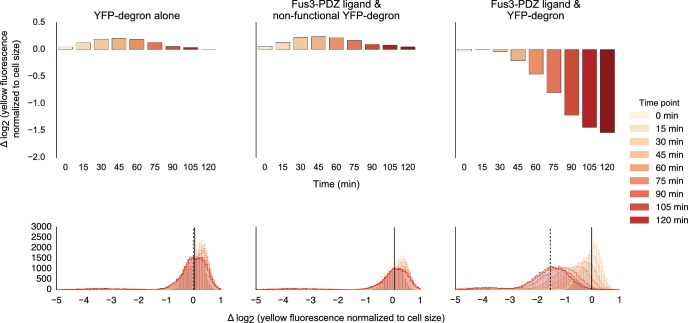
Figure 1. Rewiring the mating cascade MAPK, Fus3, to regulate the degradation of YFP.
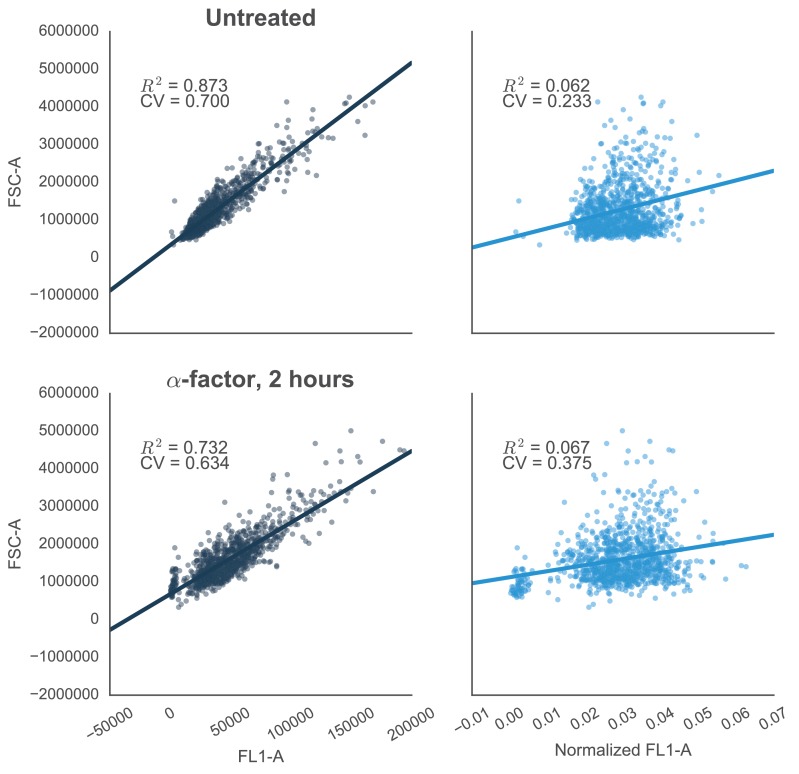
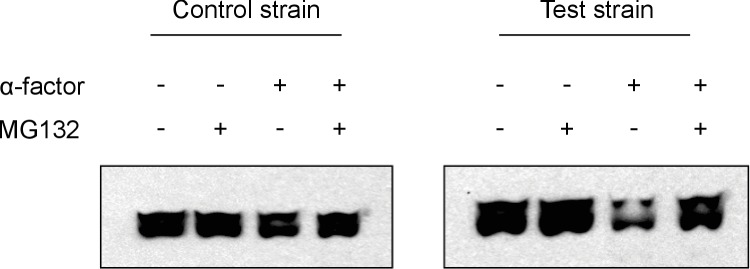
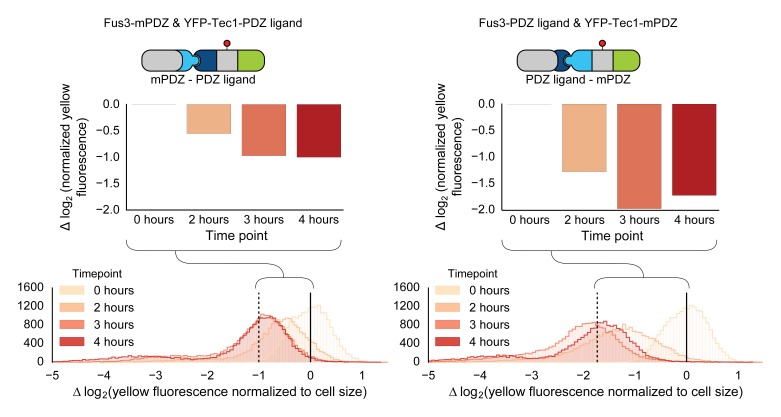
(A) The core components of the yeast mating cascade. The yeast mating factor – α-factor – triggers the sequential activation of the kinases Ste11 and Ste7 (rounded gray rectangles) followed by the MAPK, Fus3 (yellow). Arrows with red circles denote phosphorylation-mediated regulation. All three kinases are organized on the scaffold Ste5 (also gray). Among other effectors, Fus3 activates the transcription factor Ste12 (rounded gray box). (B) Fus3 targeted regulation of YFP (green). The colocalization was controlled by the addition of the mPDZ domain to YFP and a PDZ ligand to Fus3 (light blue). Degradation was mediated by the addition of a phosphodegron derived from the transcription factor Tec1 (purple). Upon activation of the mating pathway, Fus3 phosphorylates the phosphodegron fused to YFP, resulting in the recruitment of an E3 ubiquitin ligase and the ubiquitination and subsequent degradation of YFP. (C) Cells bearing the modified Fus3 and either the fully functional system, a reporter construct with an inactivated phosphodegron, a Fus3 with its kinase activity knocked out or an unmatched interaction domain (an SH3 domain instead of mPDZ) were grown to log phase and induced with 10 μM α-factor (blue histograms) or un-induced (gray histograms). Data shown are from 3 hrs post-induction. The vertical dashed black lines on the histograms represent medians of treated populations and solid black lines represent medians of untreated populations. In all figures, the fluorescence has been normalized to the cell size (see Figure 1—figure supplement 1). Full time-course experiments appear in the supplement to Figure 2.